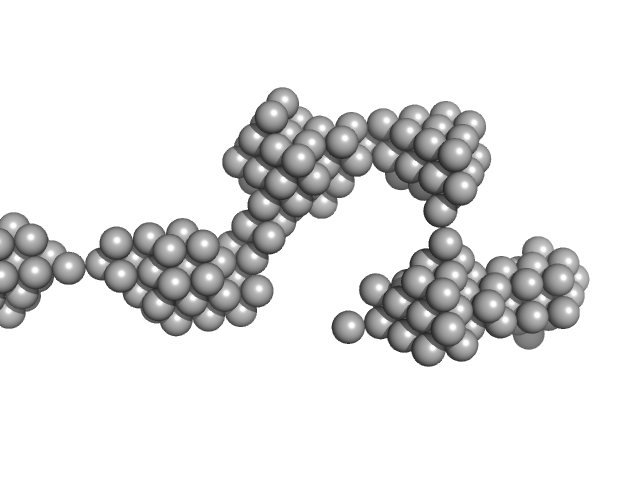
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 2
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
|
|
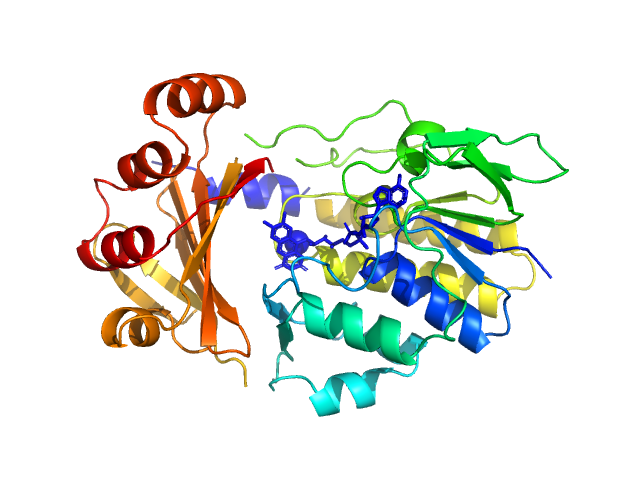
UniProt ID: A0A2Y9UCL1 (None-None) Monooxygenase (M154I, A283T)
|
|
|
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 10
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
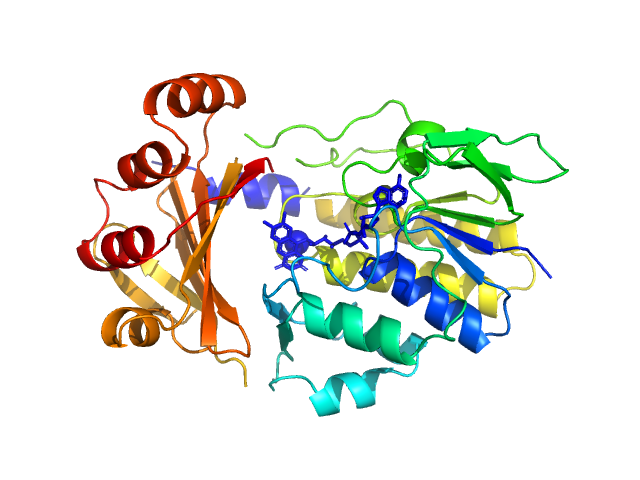
UniProt ID: A0A2Y9UCL1 (None-None) Monooxygenase (M154I, A283T)
|
|
|
|
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 30 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 7
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
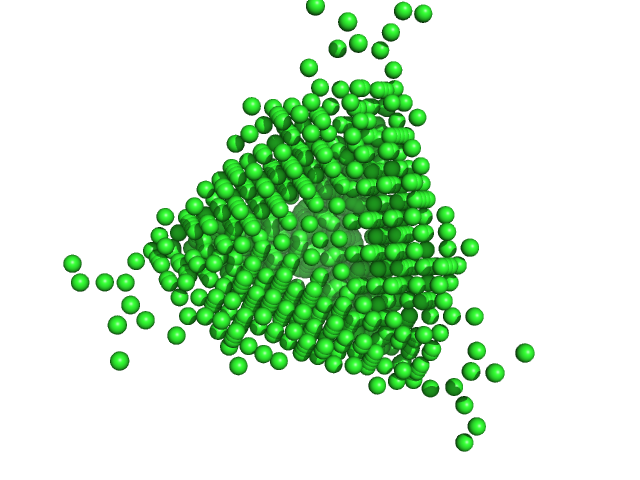
UniProt ID: P0ABT2 (1-167) DNA protection during starvation protein
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P0ABT2 (1-167) DNA protection during starvation protein
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.4 |
nm |
| Dmax |
12.0 |
nm |
|
|
UniProt ID: P0ABT2 (1-167) DNA protection during starvation protein
|
|
|
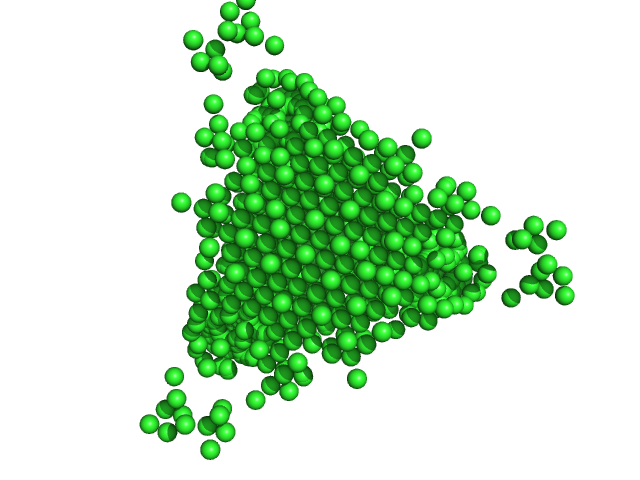
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.1 |
nm |
| Dmax |
14.0 |
nm |
|
|
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2
|
|
|
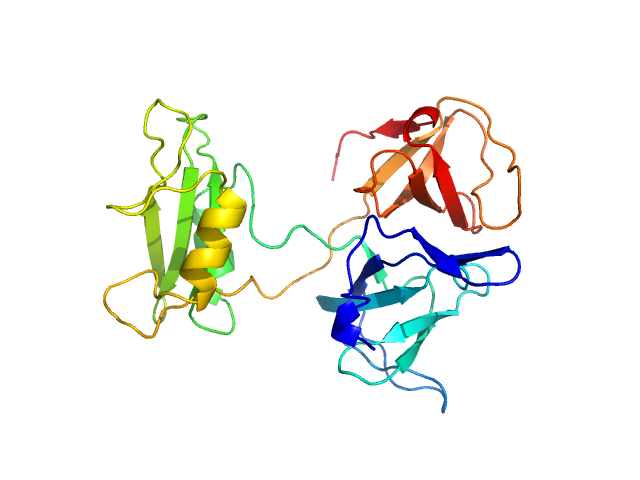
|
| Sample: |
Growth factor receptor-bound protein 2 monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2
|
|
|
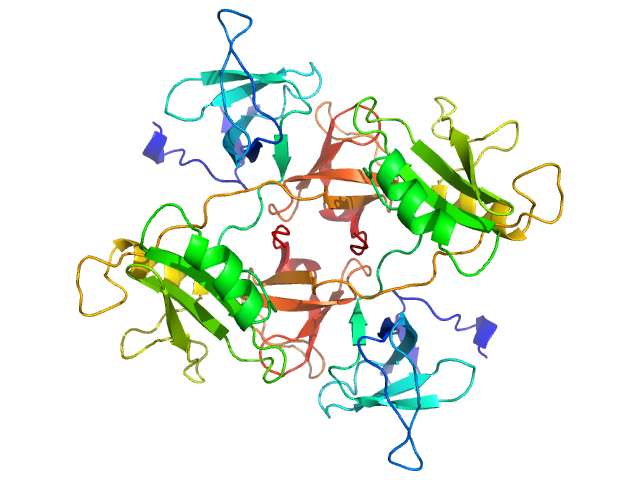
|
| Sample: |
Growth factor receptor-bound protein 2 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2 (N188D, N214D)
|
|
|
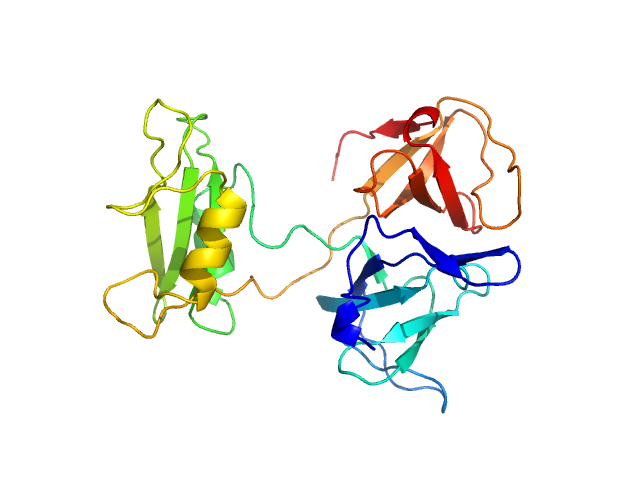
|
| Sample: |
Growth factor receptor-bound protein 2 (N188D, N214D) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2 (N188D, N214D)
|
|
|
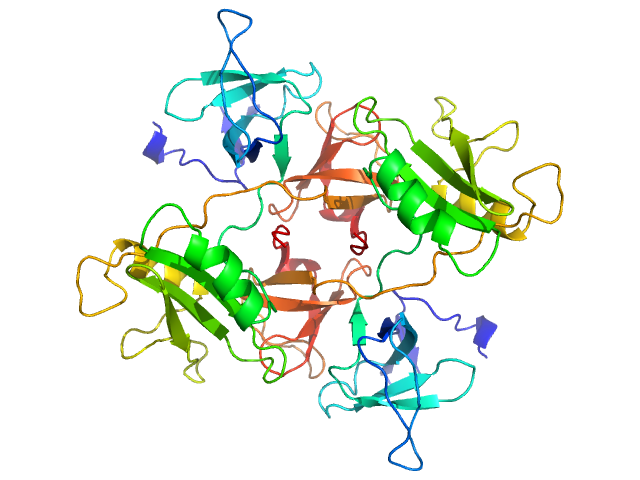
|
| Sample: |
Growth factor receptor-bound protein 2 (N188D, N214D) dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|