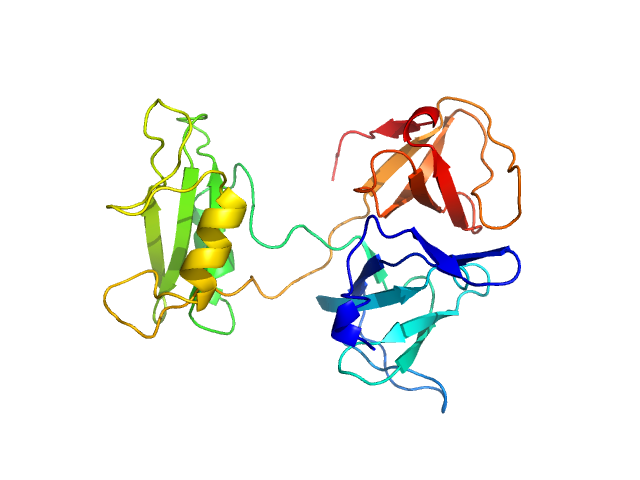
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2 (V123D)
|
|
|
|
| Sample: |
Growth factor receptor-bound protein 2 (V123D) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
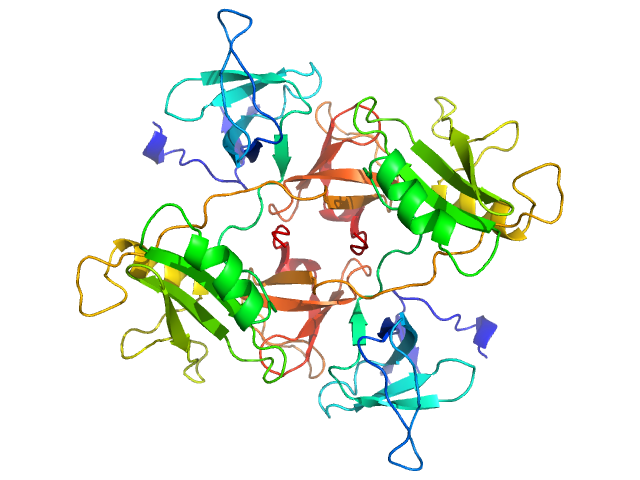
UniProt ID: P62993 (1-217) Growth factor receptor-bound protein 2 (V122P, V123P)
|
|
|
|
| Sample: |
Growth factor receptor-bound protein 2 (V122P, V123P) dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 31
|
GRB2 dimerization mediated by SH2 domain-swapping is critical for T cell signaling and cytokine production.
Sci Rep 13(1):3505 (2023)
Sandouk A, Xu Z, Baruah S, Tremblay M, Hopkins JB, Chakravarthy S, Gakhar L, Schnicker NJ, Houtman JCD
|
| RgGuinier |
3.7 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: P39935 (1-249) Eukaryotic initiation factor 4F subunit p150
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4F subunit p150, 28 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM potassium phosphate, 25 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
eIF4G1 N-terminal intrinsically disordered domain is a multi-docking station for RNA, Pab1, Pub1, and self-assembly.
Front Mol Biosci 9:986121 (2022)
Chaves-Arquero B, Martínez-Lumbreras S, Sibille N, Camero S, Bernadó P, Jiménez MÁ, Zorrilla S, Pérez-Cañadillas JM
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
137 |
nm3 |
|
|
UniProt ID: Q9BUE6 (1-129) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: Q9UMS0 (59-254) NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G)
|
|
|
|
| Sample: |
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: Q9BUE6 (1-129) Iron-sulfur cluster assembly 1 homolog, mitochondrial
UniProt ID: Q9UMS0 (59-254) NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G)
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: Q9BUE6 (1-129) Iron-sulfur cluster assembly 1 homolog, mitochondrial
UniProt ID: Q86U28 (44-154) Iron-sulfur cluster assembly 2 homolog, mitochondrial
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
Iron-sulfur cluster assembly 2 homolog, mitochondrial monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: Q9BUE6 (1-129) Iron-sulfur cluster assembly 1 homolog, mitochondrial
UniProt ID: Q9UMS0 (59-254) NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G)
UniProt ID: Q86U28 (44-154) Iron-sulfur cluster assembly 2 homolog, mitochondrial
|
|
|
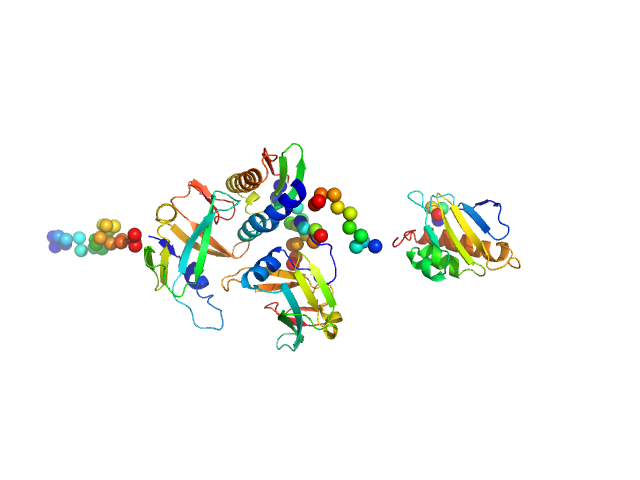
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial monomer, 14 kDa Homo sapiens protein
NFU1 iron-sulfur cluster scaffold homolog, mitochondrial (F118S, E168G) monomer, 22 kDa Homo sapiens protein
Iron-sulfur cluster assembly 2 homolog, mitochondrial monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 27
|
Structural plasticity of NFU1 upon interaction with binding partners: insights into the mitochondrial [4Fe-4S] cluster pathway
Journal of Molecular Biology :168154 (2023)
Da Vela S, Saudino G, Lucarelli F, Banci L, Svergun D, Ciofi-Baffoni S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P51531 (1373-1511) Probable global transcription activator SNF2L2 (isoform 1)
UniProt ID: P40337 (54-213) von Hippel-Lindau disease tumor suppressor
UniProt ID: Q15370 (1-104) Elongin-B
UniProt ID: Q15369 (17-112) Elongin-C
UniProt ID: None (None-None) ACBI1 protac
|
|
|
|
| Sample: |
Probable global transcription activator SNF2L2 (isoform 1) monomer, 16 kDa Homo sapiens protein
Von Hippel-Lindau disease tumor suppressor monomer, 19 kDa Homo sapiens protein
Elongin-B monomer, 12 kDa Homo sapiens protein
Elongin-C monomer, 11 kDa Homo sapiens protein
ACBI1 protac monomer, 1 kDa
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Aug 11
|
Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry.
Nat Commun 13(1):5884 (2022)
Dixon T, MacPherson D, Mostofian B, Dauzhenka T, Lotz S, McGee D, Shechter S, Shrestha UR, Wiewiora R, McDargh ZA, Pei F, Pal R, Ribeiro JV, Wilkerson T, Sachdeva V, Gao N, Jain S, Sparks S, Li Y, Vinitsky A, Zhang X, Razavi AM, Kolossváry I, Imbriglio J, Evdokimov A, Bergeron L, Zhou W, Adhikari J, Ruprecht B, Dickson A, Xu H, Sherman W, Izaguirre JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
UniProt ID: P40337 (54-213) von Hippel-Lindau disease tumor suppressor
UniProt ID: Q15370 (1-104) Elongin-B
UniProt ID: Q15369 (17-112) Elongin-C
UniProt ID: None (None-None) ACBI1 protac
UniProt ID: P51531 (1373-1511) Probable global transcription activator SNF2L2 (isoform 2)
|
|
|
|
| Sample: |
Von Hippel-Lindau disease tumor suppressor monomer, 19 kDa Homo sapiens protein
Elongin-B monomer, 12 kDa Homo sapiens protein
Elongin-C monomer, 11 kDa Homo sapiens protein
ACBI1 protac monomer, 1 kDa
Probable global transcription activator SNF2L2 (isoform 2) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Aug 11
|
Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry.
Nat Commun 13(1):5884 (2022)
Dixon T, MacPherson D, Mostofian B, Dauzhenka T, Lotz S, McGee D, Shechter S, Shrestha UR, Wiewiora R, McDargh ZA, Pei F, Pal R, Ribeiro JV, Wilkerson T, Sachdeva V, Gao N, Jain S, Sparks S, Li Y, Vinitsky A, Zhang X, Razavi AM, Kolossváry I, Imbriglio J, Evdokimov A, Bergeron L, Zhou W, Adhikari J, Ruprecht B, Dickson A, Xu H, Sherman W, Izaguirre JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
75 |
nm3 |
|
|