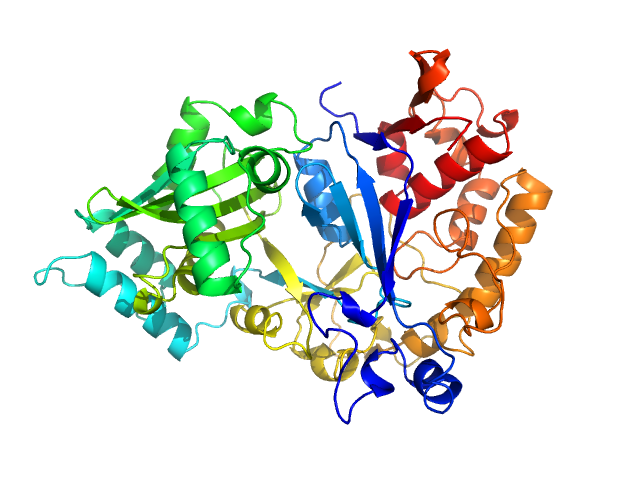
UniProt ID: P16639 (1-503) Arginyl-tRNA--protein transferase 1
|
|
|
|
| Sample: |
Arginyl-tRNA--protein transferase 1 monomer, 60 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
2% glycerol, 100 mM KCl, 50 mM Tris-HCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 12
|
The Structure of Saccharomyces cerevisiae Arginyltransferase 1 (ATE1).
J Mol Biol 434(21):167816 (2022)
Van V, Ejimogu NE, Bui TS, Smith AT
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
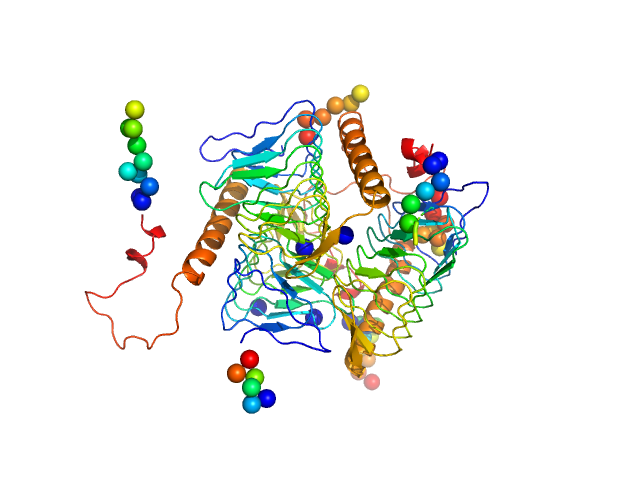
UniProt ID: D0C8J4 (1-201) Bacterial transferase hexapeptide repeat protein
|
|
|
|
| Sample: |
Bacterial transferase hexapeptide repeat protein trimer, 66 kDa Acinetobacter baumannii (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jan 29
|
Mechanistic and structural insights into the bifunctional enzyme PaaY from Acinetobacter baumannii.
Structure (2023)
Jiao M, He W, Ouyang Z, Qin Q, Guo Y, Zhang J, Bai Y, Guo X, Yu Q, She J, Hwang PM, Zheng F, Wen Y
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: Q13111 (342-475) Chromatin assembly factor 1 subunit A
|
|
|

|
| Sample: |
Chromatin assembly factor 1 subunit A monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 200 mM NaCl, 0.1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Oct 28
|
Unorthodox PCNA Binding by Chromatin Assembly Factor 1
International Journal of Molecular Sciences 23(19):11099 (2022)
Gopinathan Nair A, Rabas N, Lejon S, Homiski C, Osborne M, Cyr N, Sverzhinsky A, Melendy T, Pascal J, Laue E, Borden K, Omichinski J, Verreault A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: C9ZJ67 (18-363) 65 kDa invariant surface glycoprotein, putative
|
|
|
|
| Sample: |
65 kDa invariant surface glycoprotein, putative monomer, 41 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Nat Commun 14(1):2403 (2023)
Sülzen H, Began J, Dhillon A, Kereïche S, Pompach P, Votrubova J, Zahedifard F, Šubrtova A, Šafner M, Hubalek M, Thompson M, Zoltner M, Zoll S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: C9ZJ67 (18-363) 65 kDa invariant surface glycoprotein, putative
UniProt ID: V9HWA9 (996-1287) Complement C3
|
|
|

|
| Sample: |
65 kDa invariant surface glycoprotein, putative monomer, 41 kDa Trypanosoma brucei gambiense … protein
Complement C3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Nat Commun 14(1):2403 (2023)
Sülzen H, Began J, Dhillon A, Kereïche S, Pompach P, Votrubova J, Zahedifard F, Šubrtova A, Šafner M, Hubalek M, Thompson M, Zoltner M, Zoll S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
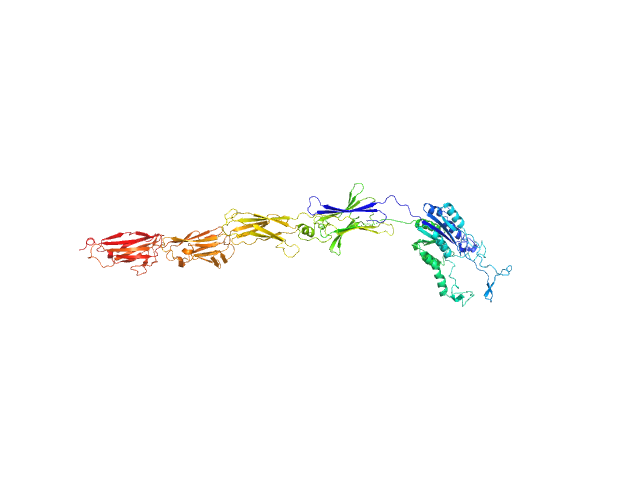
UniProt ID: D4FSQ3 (36-828) von Willebrand factor type A domain protein
|
|
|
|
| Sample: |
Von Willebrand factor type A domain protein monomer, 89 kDa Streptococcus oralis ATCC … protein
|
| Buffer: |
20 mM HEPES,150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 12
|
New structural insights into the PI-2 pilus from Streptococcus oralis, an early dental plaque colonizer.
FEBS J (2022)
Yadav RK, Krishnan V
|
| RgGuinier |
6.4 |
nm |
| Dmax |
29.2 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
UniProt ID: Q9IPJ8-3 (1-244) Isoform P3 of Phosphoprotein
|
|
|
|
| Sample: |
Isoform P3 of Phosphoprotein dimer, 55 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 26
|
Structural insights into the multifunctionality of rabies virus P3 protein.
Proc Natl Acad Sci U S A 120(14):e2217066120 (2023)
Sethi A, Rawlinson SM, Dubey A, Ang CS, Choi YH, Yan F, Okada K, Rozario AM, Brice AM, Ito N, Williamson NA, Hatters DM, Bell TDM, Arthanari H, Moseley GW, Gooley PR
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
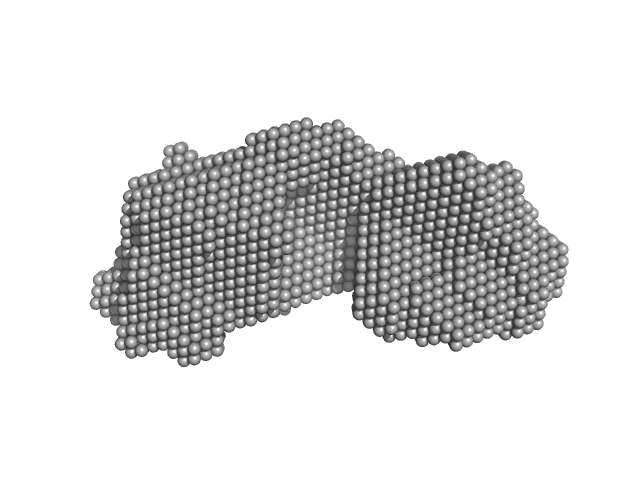
UniProt ID: B2V8L9 (2-496) SAVED domain-containing protein
|
|
|
|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 14
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
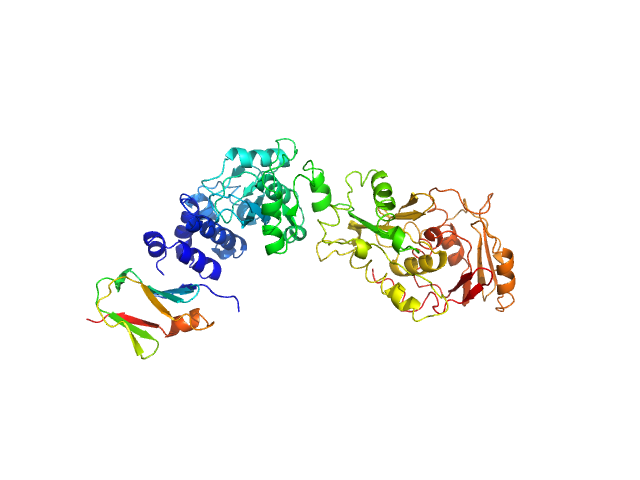
UniProt ID: B2V8L9 (2-496) SAVED domain-containing protein
UniProt ID: B2V8L8 (196-271) Uncharacterized protein (putative MazF-like toxin)
|
|
|
|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
Uncharacterized protein (putative MazF-like toxin) monomer, 9 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 14
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: B2V8L9 (2-496) SAVED domain-containing protein
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
82 |
nm3 |
|
|