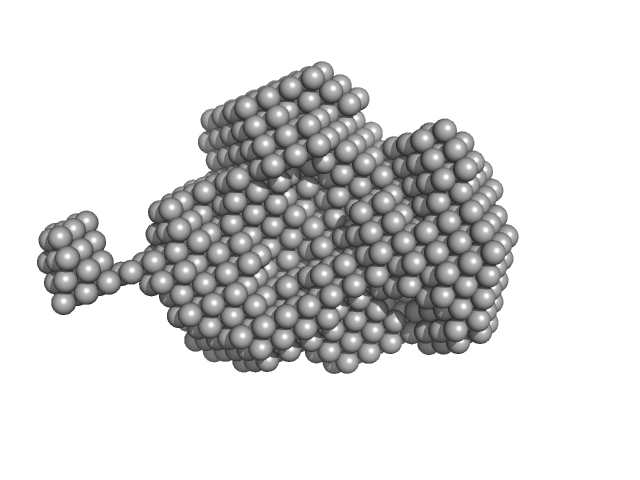
UniProt ID: P96379 (1-185) Nucleoside triphosphate pyrophosphohydrolase
|
|
|
|
| Sample: |
Nucleoside triphosphate pyrophosphohydrolase dimer, 41 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2020 Dec 11
|
Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis
Frontiers in Microbiology 14 (2023)
Wang S, Gao B, Chen A, Zhang Z, Wang S, Lv L, Zhao G, Li J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
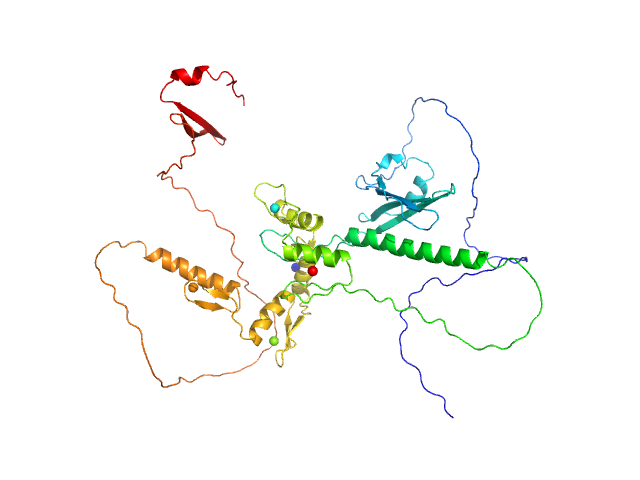
UniProt ID: Q86VK4 (1-478) Zinc finger protein 410
|
|
|
|
| Sample: |
Zinc finger protein 410 monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
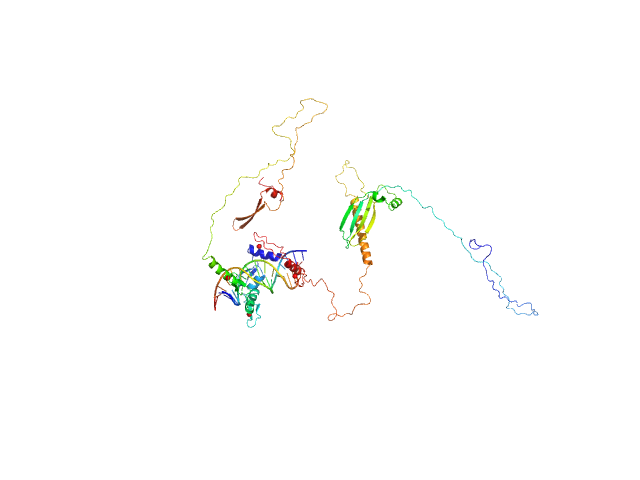
UniProt ID: Q86VK4 (1-478) Zinc finger protein 410
UniProt ID: None (None-None) DNA (Zinc finger protein 410 recognition sequence)
|
|
|
|
| Sample: |
Zinc finger protein 410 monomer, 52 kDa Homo sapiens protein
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
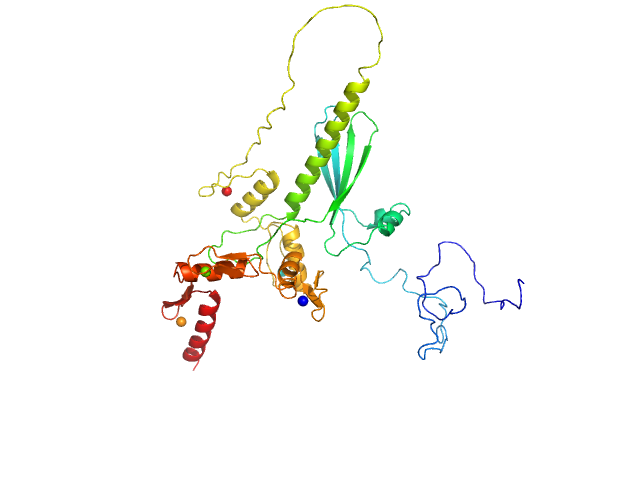
UniProt ID: Q86VK4 (1-366) Zinc finger protein 410
|
|
|
|
| Sample: |
Zinc finger protein 410 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
UniProt ID: None (None-None) DNA (Zinc finger protein 410 recognition sequence)
UniProt ID: Q86VK4 (1-366) Zinc finger protein 410
|
|
|
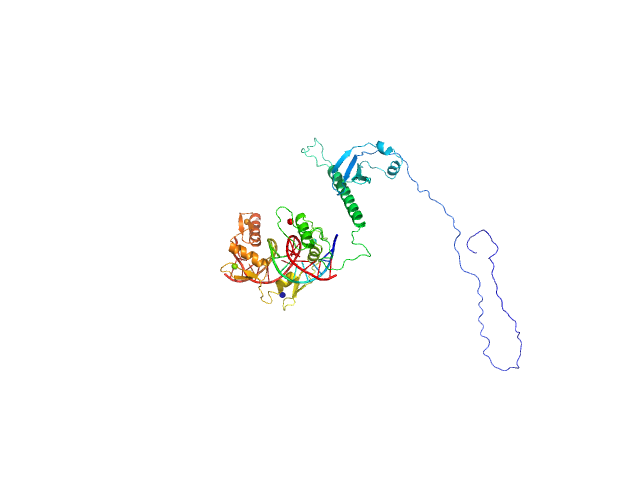
|
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
Zinc finger protein 410 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Apr 25
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
UniProt ID: Q86VK4 (217-478) Zinc finger protein 410
|
|
|
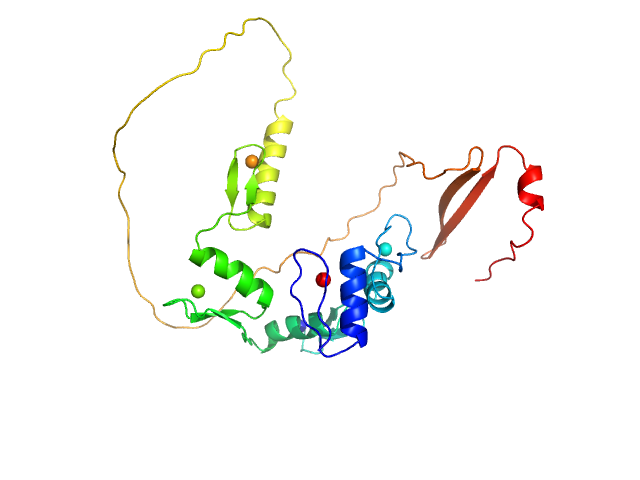
|
| Sample: |
Zinc finger protein 410 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: None (None-None) DNA (Zinc finger protein 410 recognition sequence)
UniProt ID: Q86VK4 (217-478) Zinc finger protein 410
|
|
|
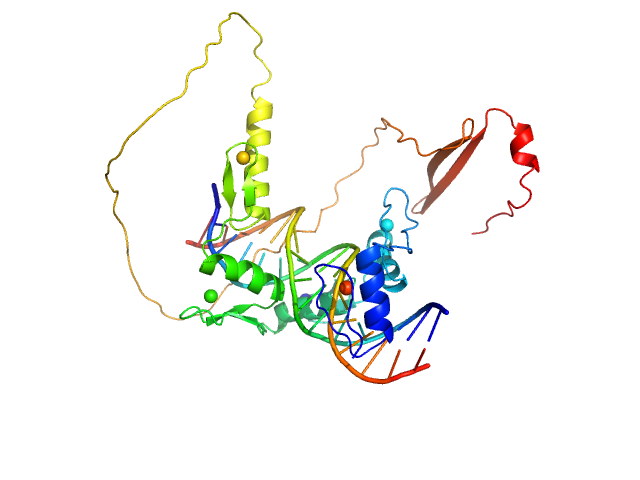
|
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
Zinc finger protein 410 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: Q6FPX9 (1-392) Metacaspase-1
|
|
|
|
| Sample: |
Metacaspase-1 monomer, 46 kDa Candida glabrata (strain … protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1% glycerol, 10 mM CaCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 26
|
Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol 5(1):1158 (2022)
Conchou L, Doumèche B, Galisson F, Violot S, Dugelay C, Diesis E, Page A, Bienvenu AL, Picot S, Aghajari N, Ballut L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: P0C799 (1-201) Phosphoprotein
|
|
|
|
| Sample: |
Phosphoprotein tetramer, 90 kDa Borna disease virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
UniProt ID: P0C799 (65-172) Phosphoprotein
|
|
|

|
| Sample: |
Phosphoprotein tetramer, 47 kDa Borna disease virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|