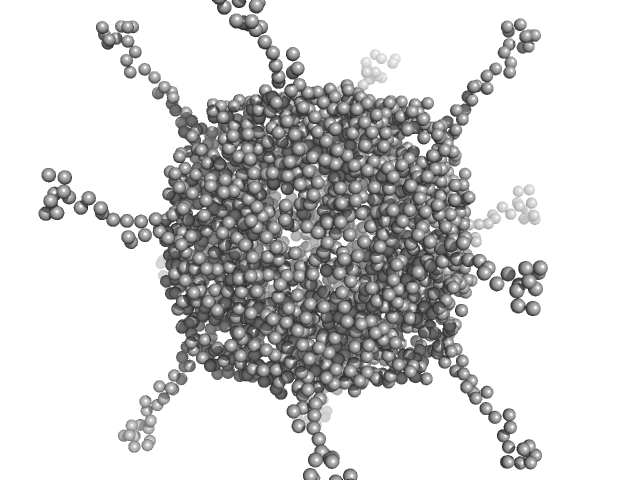
UniProt ID: A0A100HLL5 (11-214) DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation
|
|
|
|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (11-214) DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
409 |
nm3 |
|
|
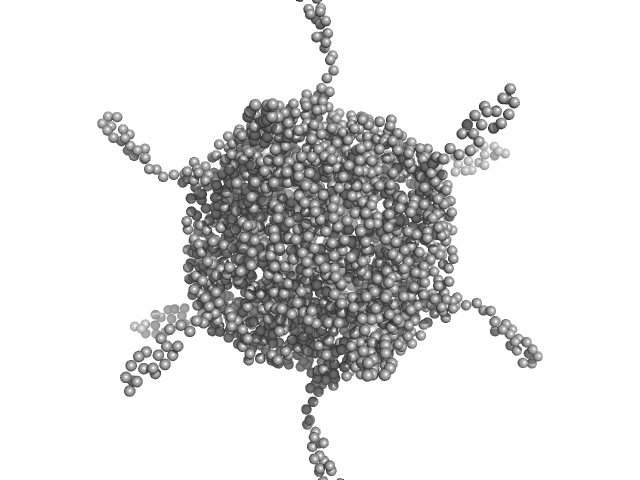
UniProt ID: A0A100HLL5 (11-214) DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation
|
|
|
|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
404 |
nm3 |
|
|
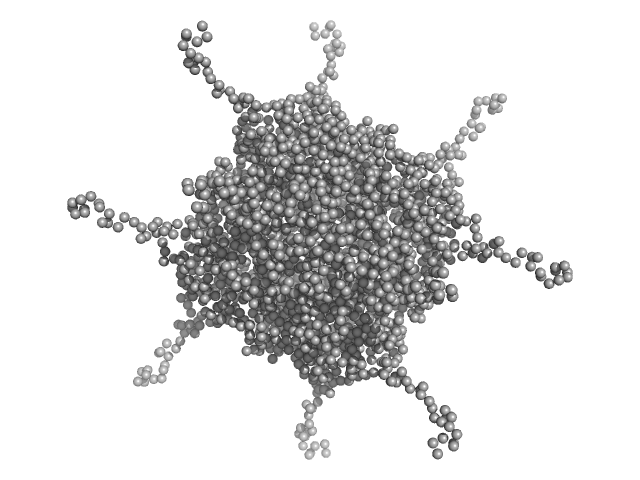
UniProt ID: A0A100HLL5 (11-214) DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation
|
|
|
|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
431 |
nm3 |
|
|
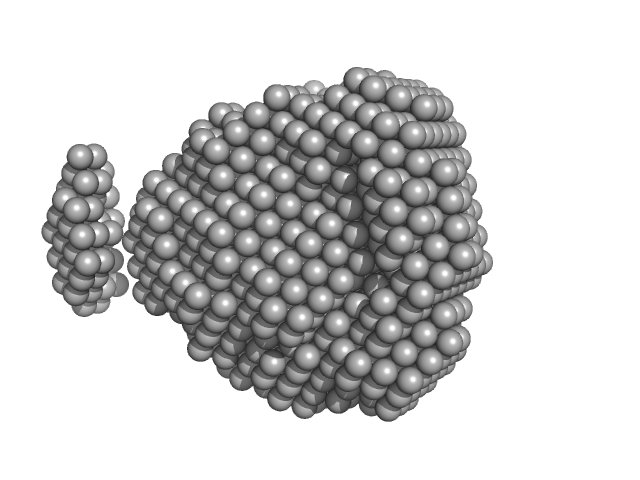
UniProt ID: B2HEF4 (1-303) Fructokinase, PfkB
|
|
|
|
| Sample: |
Fructokinase, PfkB monomer, 32 kDa Mycobacterium marinum (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2019 Dec 17
|
Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum
Biochemical and Biophysical Research Communications (2021)
Gao B, Ji R, Li Z, Su X, Li H, Sun Y, Ji C, Gan J, Li J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: P0A817 (1-384) S-adenosylmethionine synthase
UniProt ID: P07693 (1-152) S-adenosyl-L-methionine lyase
|
|
|
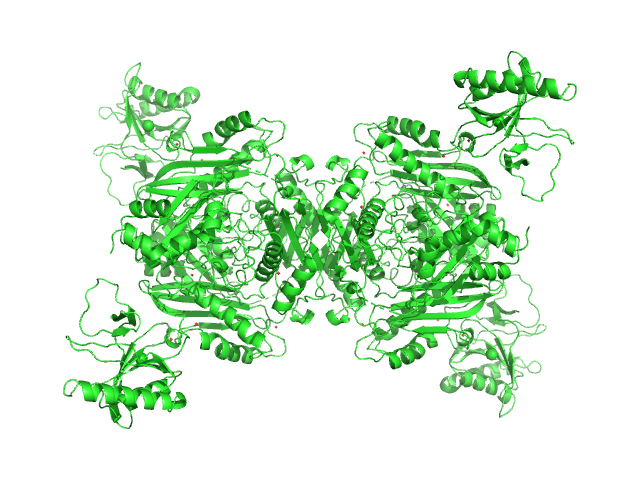
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jun 17
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
UniProt ID: P0A817 (1-384) S-adenosylmethionine synthase
UniProt ID: P07693 (None-None) S-adenosyl-L-methionine lyase (E68Q/Q94A)
|
|
|
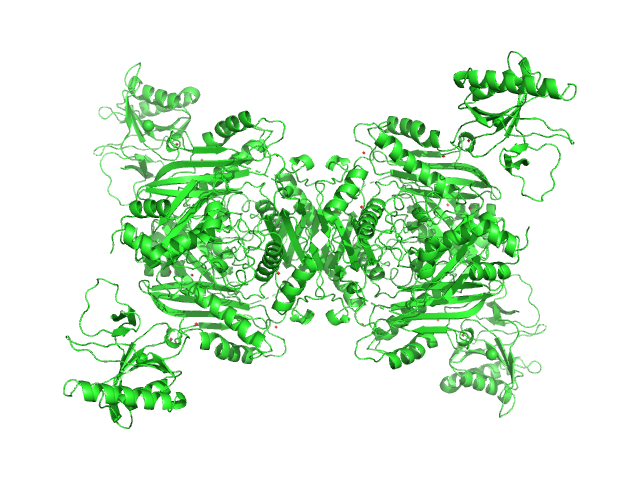
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase (E68Q/Q94A) tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 May 7
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
UniProt ID: P9WNN1 (1-396) Elongation factor Tu
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
UniProt ID: P9WNM1 (1-271) Elongation factor Ts
|
|
|

|
| Sample: |
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P9WNN1 (1-396) Elongation factor Tu
UniProt ID: P9WNM1 (None-None) Elongation factor Ts
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
111 |
nm3 |
|
|