UniProt ID: L0N429 (67-178) Phosphoprotein
|
|
|

|
| Sample: |
Phosphoprotein tetramer, 52 kDa Gaboon viper virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
UniProt ID: Q01560 (120-280) Serine/arginine (SR)-type shuttling mRNA binding protein NPL3
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q01560 (120-280) Serine/arginine (SR)-type shuttling mRNA binding protein NPL3
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: Q01560 (120-280) Serine/arginine (SR)-type shuttling mRNA binding protein NPL3
UniProt ID: None (None-None) Short RNA oligonucleotide (NPL3 binding sequence)
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
Short RNA oligonucleotide (NPL3 binding sequence) monomer, 4 kDa RNA
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
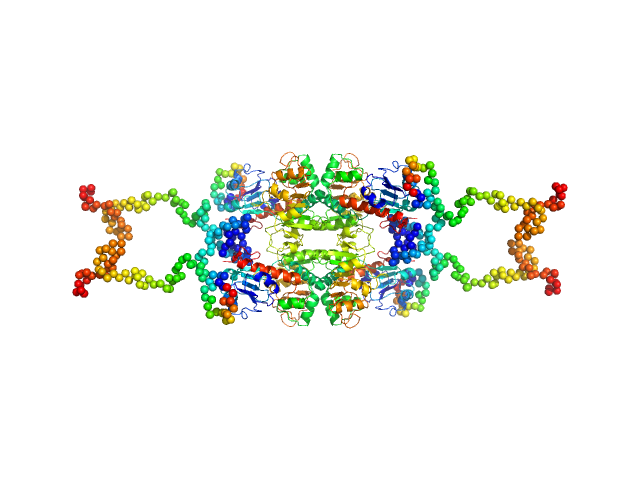
UniProt ID: Q13363 (1-440) C-terminal-binding protein 1
|
|
|
|
| Sample: |
C-terminal-binding protein 1 tetramer, 191 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
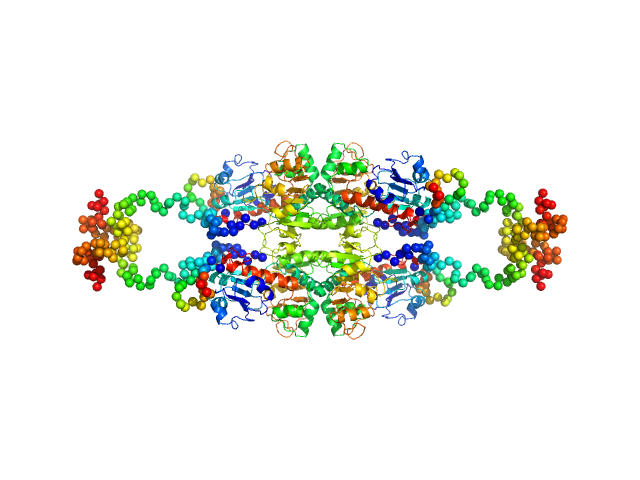
UniProt ID: Q13363 (1-440) C-terminal-binding protein 1 (C134Y, N138R, R141E, L150W)
|
|
|
|
| Sample: |
C-terminal-binding protein 1 (C134Y, N138R, R141E, L150W), 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
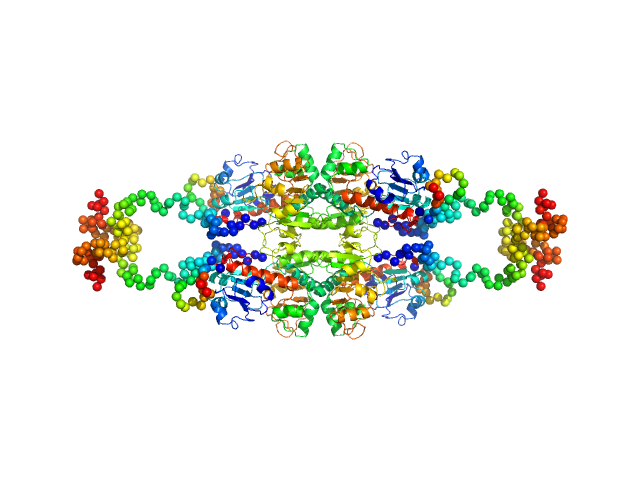
UniProt ID: Q13363 (1-440) C-terminal-binding protein 1 (R266A, D290A, E295A, H315A)
|
|
|
|
| Sample: |
C-terminal-binding protein 1 (R266A, D290A, E295A, H315A), 47 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
UniProt ID: Q9Y5P3 (303-362) Retinoic acid-induced protein 2 (303-362)
|
|
|
|
| Sample: |
Retinoic acid-induced protein 2 (303-362) monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
UniProt ID: Q9Y5P3 (303-362) Retinoic acid-induced protein 2 (303-362: L319A, S320A)
|
|
|
|
| Sample: |
Retinoic acid-induced protein 2 (303-362: L319A, S320A) monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
UniProt ID: Q9Y5P3 (303-362) Retinoic acid-induced protein 2 (303-362: L345A, S346A)
|
|
|
|
| Sample: |
Retinoic acid-induced protein 2 (303-362: L345A, S346A) monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
11 |
nm3 |
|
|