UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: None (None-None) 58 nucleotide RNA L11-binding domain from E. coli 23S rRNA
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Nov 19
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: P36238 (1-147) 50S ribosomal protein L11
|
|
|
|
| Sample: |
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: O53289 (1-409) Phosphoserine phosphatase SerB2
|
|
|
|
| Sample: |
Phosphoserine phosphatase SerB2 dimer, 87 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 6
|
A morpheein equilibrium regulates catalysis in phosphoserine phosphatase SerB2 from Mycobacterium tuberculosis.
Commun Biol 6(1):1024 (2023)
Pierson E, De Pol F, Fillet M, Wouters J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
UniProt ID: O53289 (1-409) Mycobacterium tuberculosis phosphoserine phosphatase SerB2
|
|
|
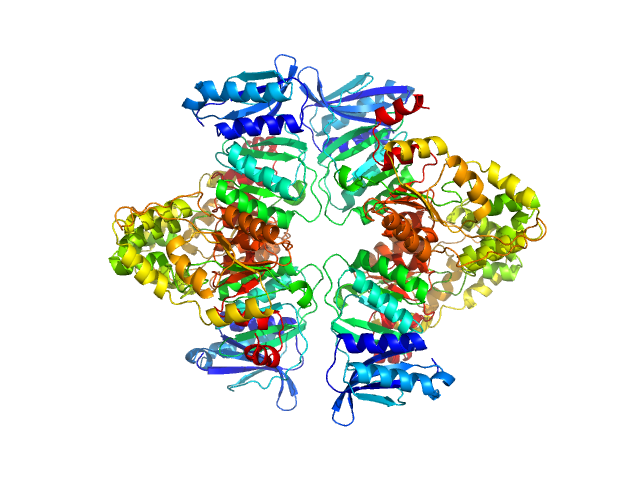
|
| Sample: |
Mycobacterium tuberculosis phosphoserine phosphatase SerB2 tetramer, 173 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 6
|
A morpheein equilibrium regulates catalysis in phosphoserine phosphatase SerB2 from Mycobacterium tuberculosis.
Commun Biol 6(1):1024 (2023)
Pierson E, De Pol F, Fillet M, Wouters J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
307 |
nm3 |
|
|