UniProt ID: F4I9G2 (96-426) Inactive purple acid phosphatase-like protein
|
|
|
|
| Sample: |
Inactive purple acid phosphatase-like protein monomer, 40 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
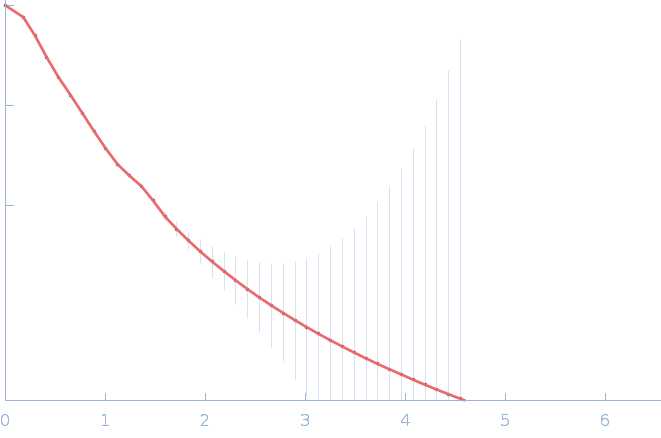
UniProt ID: P30663 (2-519) Nitrogen fixation regulatory protein (Q409L)
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
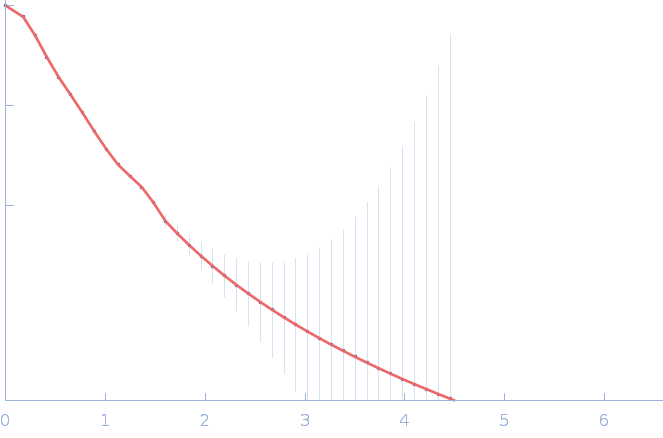
UniProt ID: P30663 (2-519) Nitrogen fixation regulatory protein (Q409L)
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
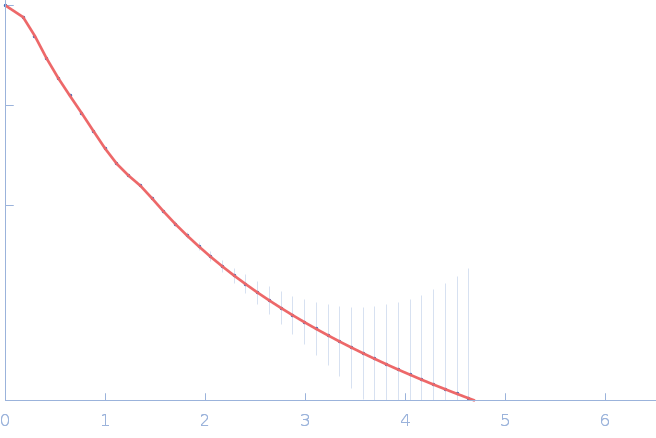
UniProt ID: P30663 (2-519) Nitrogen fixation regulatory protein (Q409L)
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|
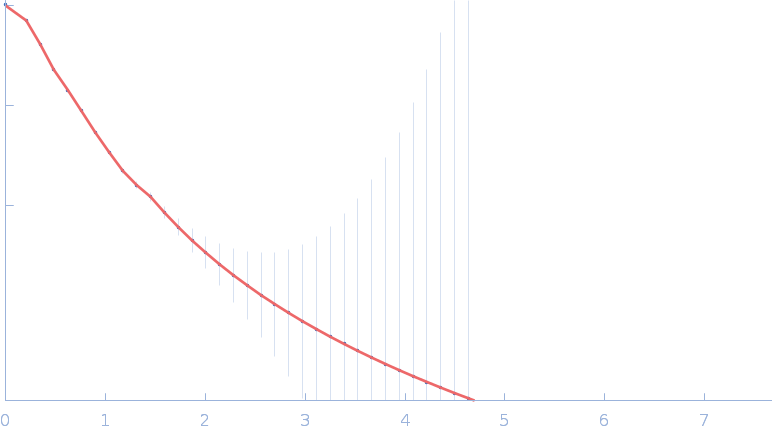
UniProt ID: P30663 (2-519) Nitrogen fixation regulatory protein (Q409L)
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
UniProt ID: Q9STE8 (141-449) Protein TOC75-3, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment tc1
|
|
|
|
| Sample: |
Protein TOC75-3, chloroplastic monomer, 35 kDa Arabidopsis thaliana protein
Synthetic antigen binding fragment tc1 monomer, 48 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
UniProt ID: Q9STE8 (141-449) Protein TOC75-3, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment tc2
|
|
|
|
| Sample: |
Protein TOC75-3, chloroplastic monomer, 35 kDa Arabidopsis thaliana protein
Synthetic antigen binding fragment tc2 monomer, 48 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
UniProt ID: Q9STE8 (141-449) Protein TOC75-3, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment tc3
|
|
|
|
| Sample: |
Protein TOC75-3, chloroplastic monomer, 35 kDa Arabidopsis thaliana protein
Synthetic antigen binding fragment tc3 monomer, 49 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
UniProt ID: Q9STE8 (141-449) Protein TOC75-3, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment tc5
|
|
|
|
| Sample: |
Protein TOC75-3, chloroplastic monomer, 35 kDa Arabidopsis thaliana protein
Synthetic antigen binding fragment tc5 monomer, 49 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
UniProt ID: Q43715 (163-440) Protein TOC75, chloroplastic
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|