UniProt ID: Q43715 (163-440) Protein TOC75, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment ax9
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
Synthetic antigen binding fragment ax9 monomer, 49 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: Q43715 (163-440) Protein TOC75, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment ax14
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
Synthetic antigen binding fragment ax14 monomer, 48 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
UniProt ID: Q43715 (163-440) Protein TOC75, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment ax15
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
Synthetic antigen binding fragment ax15 monomer, 48 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: Q43715 (163-440) Protein TOC75, chloroplastic
UniProt ID: None (None-None) synthetic antigen binding fragment ax17
|
|
|
|
| Sample: |
Protein TOC75, chloroplastic monomer, 32 kDa Pisum sativum protein
Synthetic antigen binding fragment ax17 monomer, 49 kDa synthetic construct protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 23
|
Characterization of synthetic antigen binding fragments targeting Toc75 for the isolation of TOC in A. thaliana and P. sativum
Structure (2023)
Srinivasan K, Erramilli S, Chakravarthy S, Gonzalez A, Kossiakoff A, Noinaj N
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
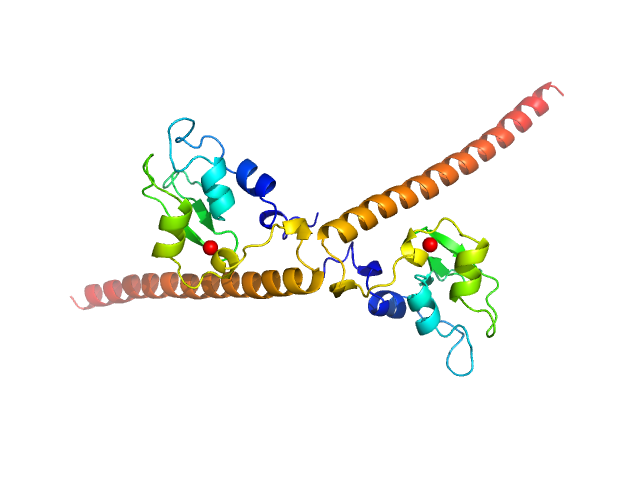
UniProt ID: O15392 (1-142) Baculoviral IAP repeat-containing protein 5
|
|
|
|
| Sample: |
Baculoviral IAP repeat-containing protein 5 dimer, 34 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 10
|
Survivin prevents the Polycomb Repressor Complex 2 from methylating Histone 3 lysine 27
iScience :106976 (2023)
Jensen M, Chandrasekaran V, García-Bonete M, Li S, Anindya A, Andersson K, Erlandsson M, Oparina N, Burmann B, Brath U, Panchenko A, Maria Bokarewa I, Katona G
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: Q8IWL3 (28-235) Iron-sulfur cluster co-chaperone protein HscB
|
|
|

|
| Sample: |
Iron-sulfur cluster co-chaperone protein HscB monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, 5 mM KCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 Aug 14
|
Structural characterization of the human DjC20/HscB cochaperone in solution
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140970 (2023)
de Souza Coto A, Pereira A, Oliveira S, de Oliveira Moritz M, da Rocha A, Dores-Silva P, da Silva N, de Araújo Nogueira A, Gava L, Seraphim T, Borges J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P9WNA1 (1-99) Cell division protein FtsQ
|
|
|
|
| Sample: |
Cell division protein FtsQ monomer, 11 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM phosphate buffer, 25 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at 5-ID-D DND-CAT, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jun 28
|
An Arg/Ala-rich helix in the N-terminal region of M. tuberculosis FtsQ is a potential membrane anchor of the Z-ring.
Commun Biol 6(1):311 (2023)
Smrt ST, Escobar CA, Dey S, Cross TA, Zhou HX
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: Q16539 (10-360) Mitogen-activated protein kinase 14
UniProt ID: None (None-None) Dual specificity mitogen-activated protein kinase kinase 6
UniProt ID: None (None-None) Adenosine 5'-Diphosphate Tetrafluoroaluminate
|
|
|
|
| Sample: |
Mitogen-activated protein kinase 14 monomer, 40 kDa Homo sapiens protein
Dual specificity mitogen-activated protein kinase kinase 6 monomer, 42 kDa synthetic construct protein
Adenosine 5'-Diphosphate Tetrafluoroaluminate monomer, 1 kDa
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 10 mM MgCl2, 5 % glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 15
|
Architecture of the MKK6-p38α complex defines the basis of MAPK specificity and activation
Science 381(6663):1217-1225 (2023)
Juyoux P, Galdadas I, Gobbo D, von Velsen J, Pelosse M, Tully M, Vadas O, Gervasio F, Pellegrini E, Bowler M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
UniProt ID: Q16539 (10-360) Mitogen-activated protein kinase 14
UniProt ID: None (None-None) Dual specificity mitogen-activated protein kinase kinase 6
UniProt ID: None (None-None) 5′‐adenylyl methylenediphosphonate
|
|
|
|
| Sample: |
Mitogen-activated protein kinase 14 monomer, 40 kDa Homo sapiens protein
Dual specificity mitogen-activated protein kinase kinase 6 monomer, 42 kDa synthetic construct protein
5′‐adenylyl methylenediphosphonate monomer, 1 kDa
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 10 mM MgCl2, 5 % glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 15
|
Architecture of the MKK6-p38α complex defines the basis of MAPK specificity and activation
Science 381(6663):1217-1225 (2023)
Juyoux P, Galdadas I, Gobbo D, von Velsen J, Pelosse M, Tully M, Vadas O, Gervasio F, Pellegrini E, Bowler M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
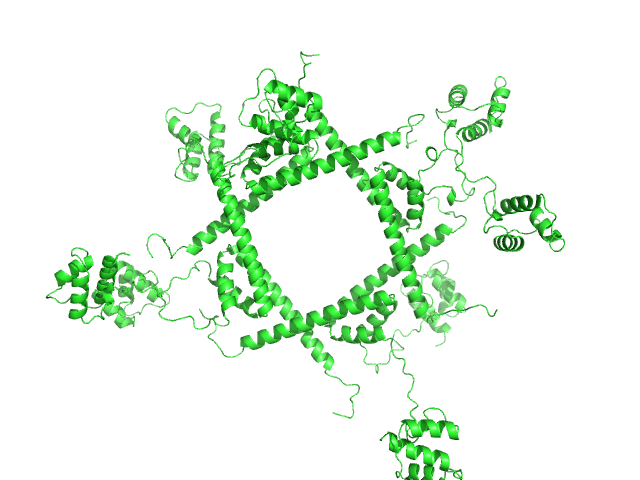
UniProt ID: Q8XAD6 (1-123) Phage repressor protein CI (C120S)
|
|
|
|
| Sample: |
Phage repressor protein CI (C120S) octamer, 122 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Regulation of the Escherichia coli paaR2-paaA2-ParD2 toxin-antitoxin system
Marusa Prolic Kalinsek
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
428 |
nm3 |
|
|