UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRU8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
39.5 |
nm |
| VolumePorod |
1127 |
nm3 |
|
|
UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRV8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
350 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
44.5 |
nm |
| VolumePorod |
1150 |
nm3 |
|
|
UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRW8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
450 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
10.8 |
nm |
| Dmax |
46.5 |
nm |
| VolumePorod |
1703 |
nm3 |
|
|
UniProt ID: Q14141 (317-427) Septin-6 (C-terminal domain of SEPT6 fused to his-tagged MBP)
UniProt ID: Q16181 (335-437) Septin-7 (C-terminal domain of SEPT7 fused to SUMO)
|
|
|
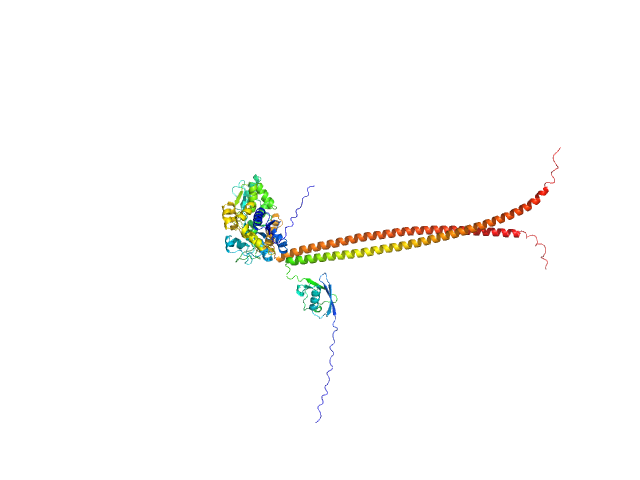
|
| Sample: |
Septin-6 (C-terminal domain of SEPT6 fused to his-tagged MBP) monomer, 55 kDa Homo sapiens protein
Septin-7 (C-terminal domain of SEPT7 fused to SUMO) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 3
|
X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol (2023)
Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
UniProt ID: Q16181 (335-437) Septin-7 (C-terminal domain of SEPT7 fused to SUMO)
UniProt ID: Q92599 (319-429) Septin-8 (C-terminal domain of SEPT8 fused to MBP)
|
|
|
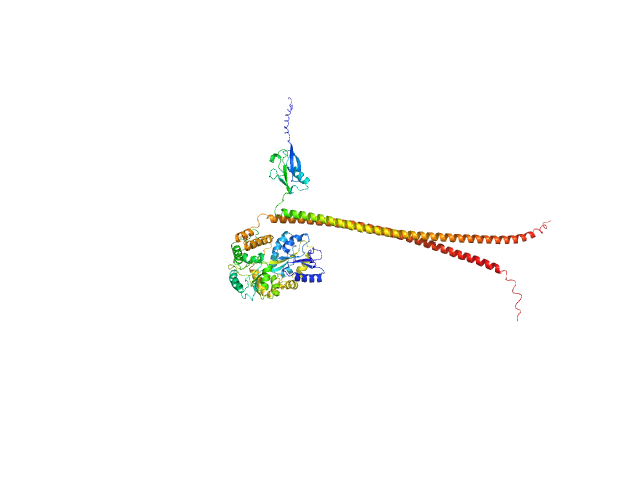
|
| Sample: |
Septin-7 (C-terminal domain of SEPT7 fused to SUMO) monomer, 25 kDa Homo sapiens protein
Septin-8 (C-terminal domain of SEPT8 fused to MBP) monomer, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 3
|
X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol (2023)
Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: Q16181 (335-437) Septin-7 (C-terminal domain of SEPT7 fused to SUMO)
UniProt ID: Q9P0V9 (341-454) Septin-10 (C-terminal domain of SEPT10 fused to MBP)
|
|
|

|
| Sample: |
Septin-7 (C-terminal domain of SEPT7 fused to SUMO) monomer, 25 kDa Homo sapiens protein
Septin-10 (C-terminal domain of SEPT10 fused to MBP) monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 3
|
X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol (2023)
Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
UniProt ID: Q16181 (335-437) Septin-7 (C-terminal domain of SEPT7 fused to SUMO)
UniProt ID: Q9NVA2 (316-429) Septin-11 (C-terminal domain of SEPT11 fused to MBP)
|
|
|
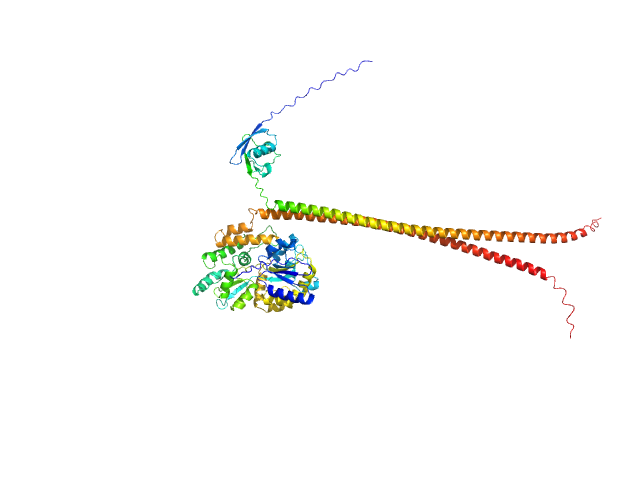
|
| Sample: |
Septin-7 (C-terminal domain of SEPT7 fused to SUMO) monomer, 25 kDa Homo sapiens protein
Septin-11 (C-terminal domain of SEPT11 fused to MBP) monomer, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 3
|
X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol (2023)
Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC
|
| RgGuinier |
5.2 |
nm |
| Dmax |
20.9 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
UniProt ID: Q16181 (335-437) Septin-7 (C-terminal domain of SEPT7 fused to SUMO)
UniProt ID: Q6ZU15 (327-432) Septin-14 (C-terminal domain of SEPT14 fused to MBP)
|
|
|
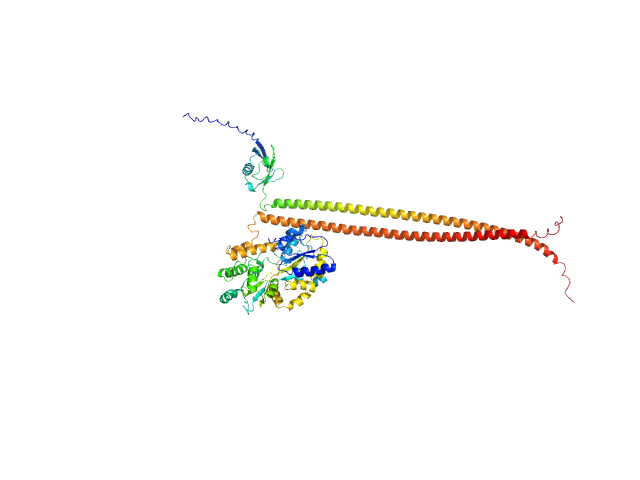
|
| Sample: |
Septin-7 (C-terminal domain of SEPT7 fused to SUMO) monomer, 25 kDa Homo sapiens protein
Septin-14 (C-terminal domain of SEPT14 fused to MBP) monomer, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 3
|
X-ray structure of the metastable SEPT14-SEPT7 coiled coil reveals a hendecad region crucial for heterodimerization.
Acta Crystallogr D Struct Biol (2023)
Cavini IA, Winter AJ, D'Muniz Pereira H, Woolfson DN, Crump MP, Garratt RC
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.3 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
UniProt ID: P20711 (None-None) Aromatic-L-amino-acid decarboxylase (M17V)
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
UniProt ID: P20711 (None-None) Aromatic-L-amino-acid decarboxylase (M17V)
|
|
|
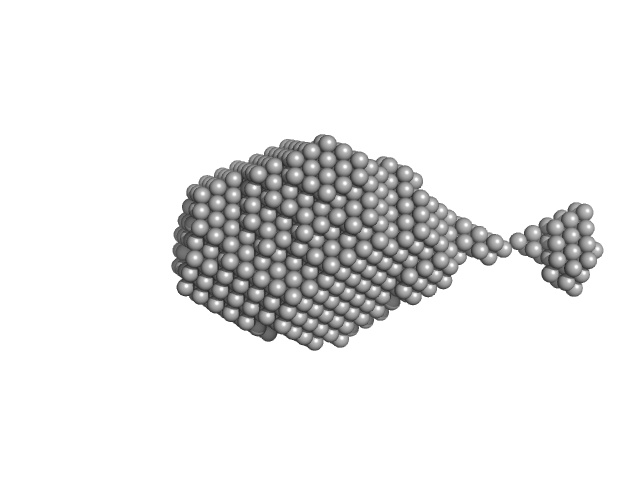
|
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|