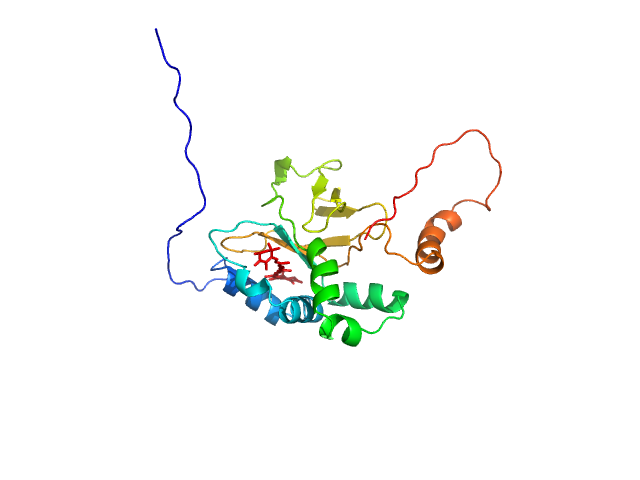
UniProt ID: S6BQ14 (21-223) Astaxanthin binding fasciclin family protein
|
|
|
|
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
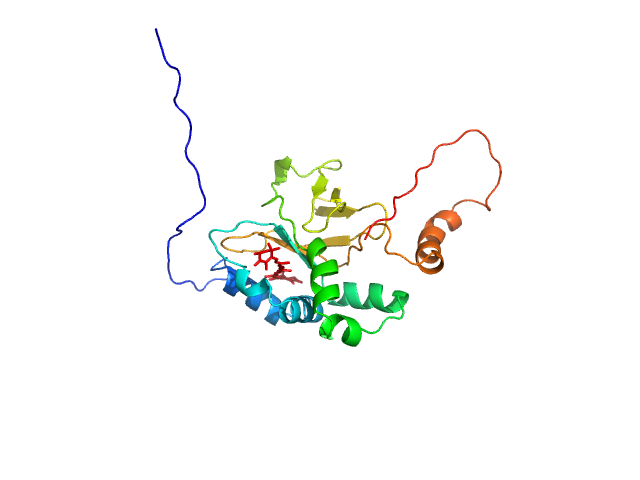
UniProt ID: S6BQ14 (21-223) Astaxanthin binding fasciclin family protein
|
|
|
|
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: None (None-None) HOTag-(PA)25-Ubiquitin
|
|
|
|
| Sample: |
HOTag-(PA)25-Ubiquitin tetramer, 68 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 22
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
UniProt ID: P02769 (25-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Jun 7
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM sodium acetate, 50 mM NaCl, pH: 4 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 2
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
UniProt ID: P69905 (1-142) Hemoglobin subunit alpha
UniProt ID: P68871 (1-147) Hemoglobin subunit beta
|
|
|
|
| Sample: |
Hemoglobin subunit alpha dimer, 31 kDa Homo sapiens protein
Hemoglobin subunit beta dimer, 32 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Oct 7
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
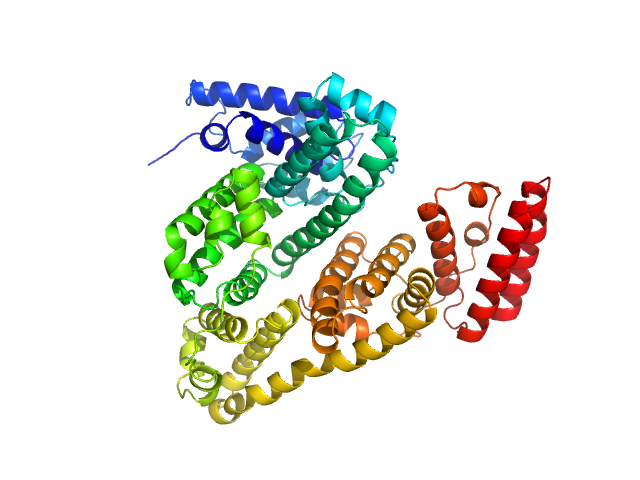
UniProt ID: P02769 (25-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 5 mM NaCl, 0.1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jun 8
|
Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering.
Sci Rep 13(1):15764 (2023)
Graewert MA, Wilhelmy C, Bacic T, Schumacher J, Blanchet C, Meier F, Drexel R, Welz R, Kolb B, Bartels K, Nawroth T, Klein T, Svergun D, Langguth P, Haas H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
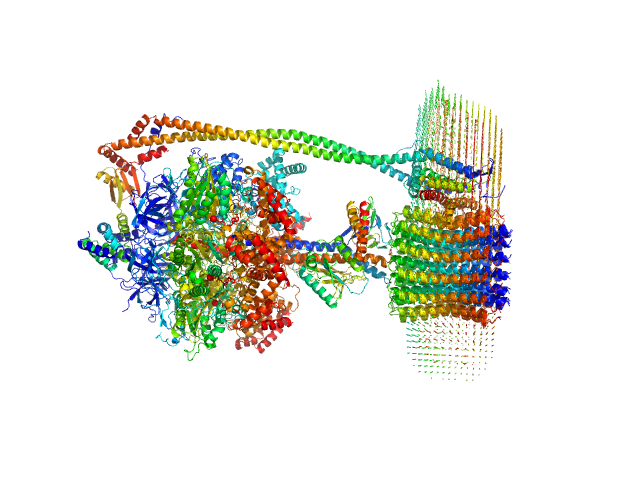
UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
300 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
6.6 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
927 |
nm3 |
|
|
UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
150 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
9.6 |
nm |
| Dmax |
41.5 |
nm |
| VolumePorod |
1506 |
nm3 |
|
|
UniProt ID: P06450 (1-507) ATP synthase subunit alpha, chloroplastic
UniProt ID: P00825 (1-498) ATP synthase subunit beta, chloroplastic
UniProt ID: P05435 (1-364) ATP synthase gamma chain, chloroplastic
UniProt ID: P11402 (1-257) ATP synthase delta chain, chloroplastic
UniProt ID: P00833 (1-134) ATP synthase epsilon chain, chloroplastic
UniProt ID: P06451 (1-247) ATP synthase subunit a, chloroplastic
UniProt ID: P06453 (1-184) ATP synthase subunit b, chloroplastic
UniProt ID: P31853 (1-222) ATP synthase subunit b', chloroplastic
UniProt ID: P69447 (1-81) ATP synthase subunit c, chloroplastic
UniProt ID: None (None-None) 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT8_fit1_model1.png)
|
| Sample: |
ATP synthase subunit alpha, chloroplastic trimer, 166 kDa Spinacia oleracea protein
ATP synthase subunit beta, chloroplastic trimer, 161 kDa Spinacia oleracea protein
ATP synthase gamma chain, chloroplastic monomer, 40 kDa Spinacia oleracea protein
ATP synthase delta chain, chloroplastic monomer, 28 kDa Spinacia oleracea protein
ATP synthase epsilon chain, chloroplastic monomer, 15 kDa Spinacia oleracea protein
ATP synthase subunit a, chloroplastic monomer, 27 kDa Spinacia oleracea protein
ATP synthase subunit b, chloroplastic monomer, 21 kDa Spinacia oleracea protein
ATP synthase subunit b', chloroplastic monomer, 24 kDa Spinacia oleracea protein
ATP synthase subunit c, chloroplastic 14-mer, 112 kDa Spinacia oleracea protein
4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside 0, 283 kDa
|
| Buffer: |
250 mM NaCl, 30 mM HEPES, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Oct 3
|
I-Shaped Dimers of a Plant Chloroplast FOF1-ATP Synthase in Response to Changes in Ionic Strength
International Journal of Molecular Sciences 24(13):10720 (2023)
Osipov S, Ryzhykau Y, Zinovev E, Minaeva A, Ivashchenko S, Verteletskiy D, Sudarev V, Kuklina D, Nikolaev M, Semenov Y, Zagryadskaya Y, Okhrimenko I, Gette M, Dronova E, Shishkin A, Dencher N, Kuklin A, Ivanovich V, Uversky V, Vlasov A
|
| RgGuinier |
7.4 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
949 |
nm3 |
|
|