UniProt ID: P20711 (None-None) Aromatic-L-amino-acid decarboxylase (M17V)
|
|
|
|
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
UniProt ID: P35991 (1-659) Tyrosine-protein kinase BTK
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK monomer, 77 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
4.1 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
UniProt ID: P35991 (1-659) Tyrosine-protein kinase BTK (A384P, S386P, T387P, A388P, L390F)
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (A384P, S386P, T387P, A388P, L390F) monomer, 77 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
3.9 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
UniProt ID: P35991 (214-659) Tyrosine-protein kinase BTK (SH3-SH2-kinase domains)
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (SH3-SH2-kinase domains) monomer, 52 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: P35991 (214-659) Tyrosine-protein kinase BTK (SH3-SH2-kinase domain A384P, S386P, T387P, A388P, L390F)
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (SH3-SH2-kinase domain A384P, S386P, T387P, A388P, L390F) monomer, 53 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 2% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 9
|
Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife 12 (2024)
Lin DY, Kueffer LE, Juneja P, Wales TE, Engen JR, Andreotti AH
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
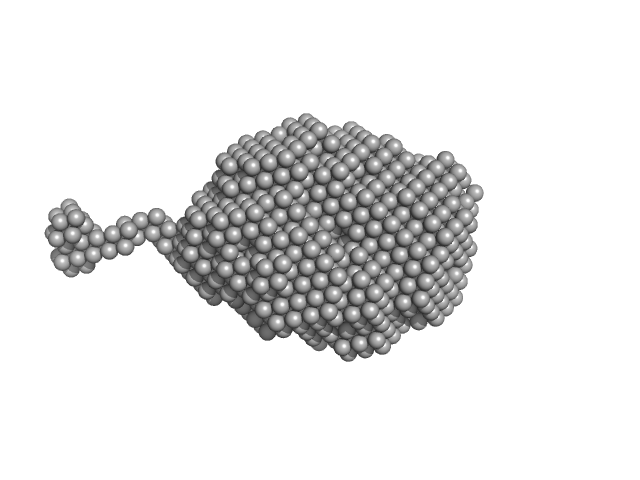
UniProt ID: P48061 (22-89) Stromal cell-derived factor 1
|
|
|
|
| Sample: |
Stromal cell-derived factor 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
UniProt ID: P63159 (2-215) High mobility group protein B1 (D189E, E202D, E215D)
|
|
|
|
| Sample: |
High mobility group protein B1 (D189E, E202D, E215D) monomer, 25 kDa Rattus norvegicus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: P48061 (22-89) Stromal cell-derived factor 1
UniProt ID: P63159 (2-215) High mobility group protein B1 (D189E, E202D, E215D)
|
|
|
|
| Sample: |
Stromal cell-derived factor 1 monomer, 9 kDa Homo sapiens protein
High mobility group protein B1 (D189E, E202D, E215D) monomer, 25 kDa Rattus norvegicus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
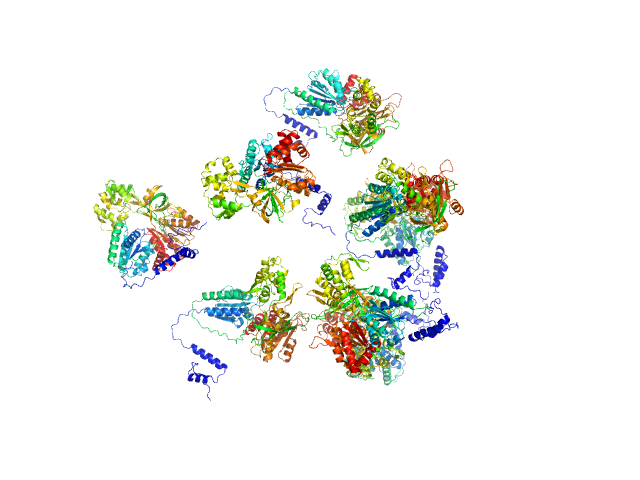
UniProt ID: P38038 (1-599) Sulfite reductase [NADPH] flavoprotein alpha-component
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component octamer, 530 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jun 24
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
8.4 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
767 |
nm3 |
|
|
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component
UniProt ID: P38038 (1-599) Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2023 Apr 24
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
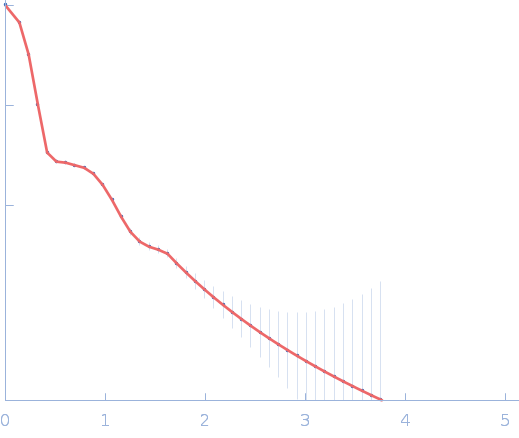
| RgGuinier |
7.5 |
nm |
| Dmax |
22.6 |
nm |
| VolumePorod |
823 |
nm3 |
|
|