UniProt ID: A0A2S5LZS0 (3-314) Sensor domain-containing diguanylate cyclase
|
|
|
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: P03372 (1-184) Estrogen receptor (mutant S118A)
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118A) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
UniProt ID: P03372 (1-184) Estrogen receptor (mutant S118D)
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118D) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
UniProt ID: Q02817 (1781-1892) Mucin-2 (CysD2 domain)
|
|
|
|
| Sample: |
Mucin-2 (CysD2 domain) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 70 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 8
|
The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep 43(5):114207 (2024)
Recktenwald CV, Karlsson G, Garcia-Bonete MJ, Katona G, Jensen M, Lymer R, Bäckström M, Johansson MEV, Hansson GC, Trillo-Muyo S
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
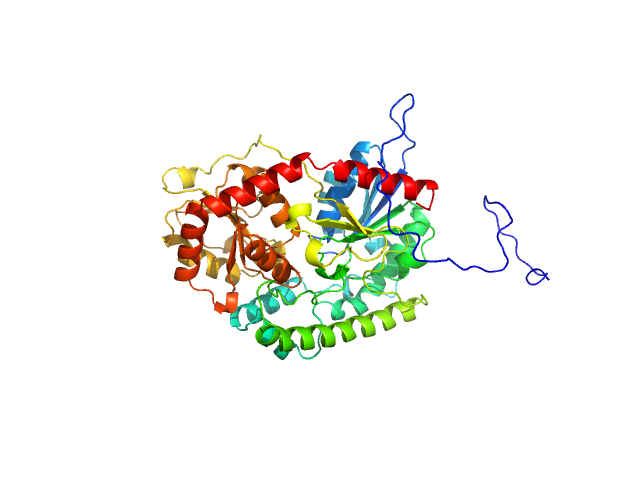
UniProt ID: T1KUK4 (1-437) UDP-glycosyltransferase 202A2
|
|
|
|
| Sample: |
UDP-glycosyltransferase 202A2 monomer, 53 kDa Tetranychus urticae protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 17
|
Structural and functional studies reveal the molecular basis of substrate promiscuity of a glycosyltransferase originating from a major agricultural pest
Journal of Biological Chemistry :105421 (2023)
Arriaza R, Abiskaroon B, Patel M, Daneshian L, Kluza A, Snoeck S, Watkins M, Hopkins J, Van Leeuwen T, Grbic M, Grbic V, Borowski T, Chruszcz M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: P0DTD1 (3264-3569) Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
UniProt ID: P0DTD1 (3264-3569) Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: P0DTD1 (3264-3569) Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
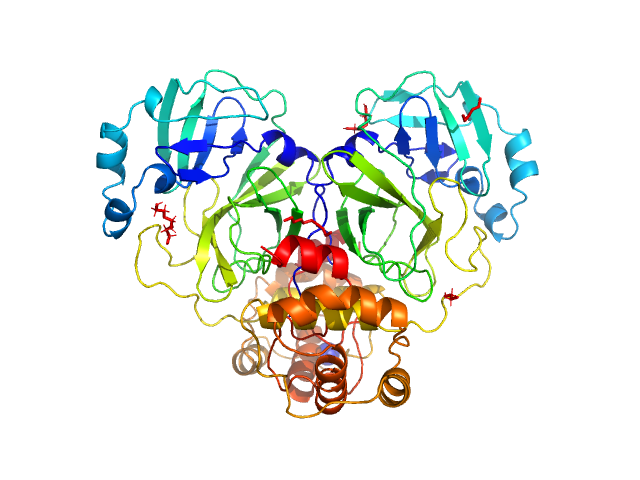
UniProt ID: P0DTD1 (3264-3569) Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: P0DTD1 (3264-3569) Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
98 |
nm3 |
|
|