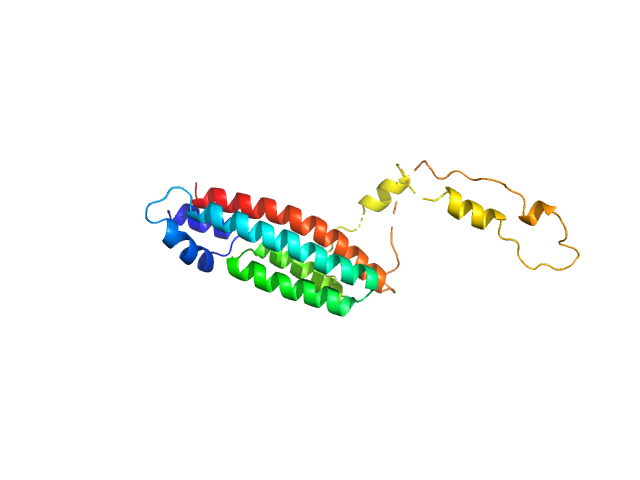
UniProt ID: Q59WC6 (12-184) Phosphorelay intermediate protein YPD1
|
|
|
|
| Sample: |
Phosphorelay intermediate protein YPD1 monomer, 19 kDa Candida albicans (strain … protein
|
| Buffer: |
50 mM Tris-HCl pH 8, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Oct 10
|
Structural and functional insights underlying recognition of histidine phosphotransfer protein in fungal phosphorelay systems
Communications Biology 7(1) (2024)
Paredes-Martínez F, Eixerés L, Zamora-Caballero S, Casino P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: D1MZ11 (49-197) Ubiquinol-cytochrome c reductase iron-sulfur subunit
|
|
|
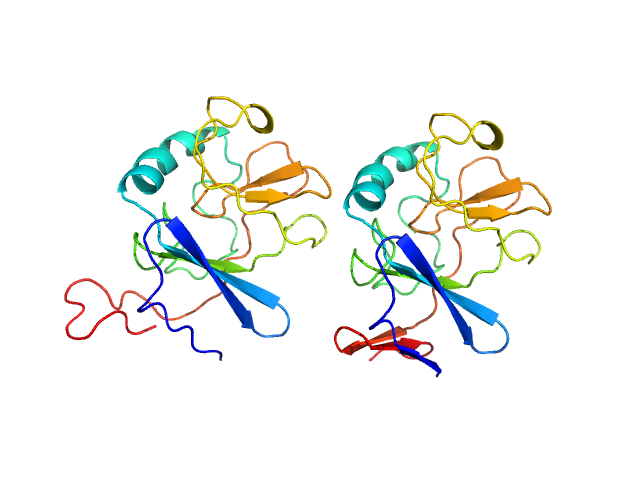
|
| Sample: |
Ubiquinol-cytochrome c reductase iron-sulfur subunit dimer, 33 kDa Thermochromatium tepidum protein
|
| Buffer: |
10 mM Tris-HCl pH 7.6, 150 mM NaCl, 5% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2023 Apr 26
|
Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.
Commun Chem 6(1):190 (2023)
Tsutsumi E, Niwa S, Takeda R, Sakamoto N, Okatsu K, Fukai S, Ago H, Nagao S, Sekiguchi H, Takeda K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: Q4VC05-2 (1-231) Isoform 2 of B-cell CLL/lymphoma 7 protein family member A
|
|
|
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.8 |
nm |
| Dmax |
28.8 |
nm |
|
|
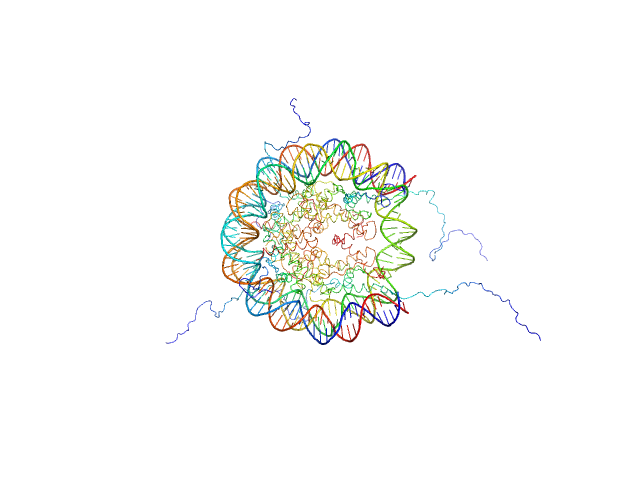
UniProt ID: P84233 (1-135) Histone H3.2
UniProt ID: P62799 (1-103) Histone H4
UniProt ID: Q6AZJ8 (1-130) Histone H2A
UniProt ID: P02281 (1-126) Histone H2B 1.1
UniProt ID: None (None-None) Widom 601 DNA sequence
|
|
|
|
| Sample: |
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
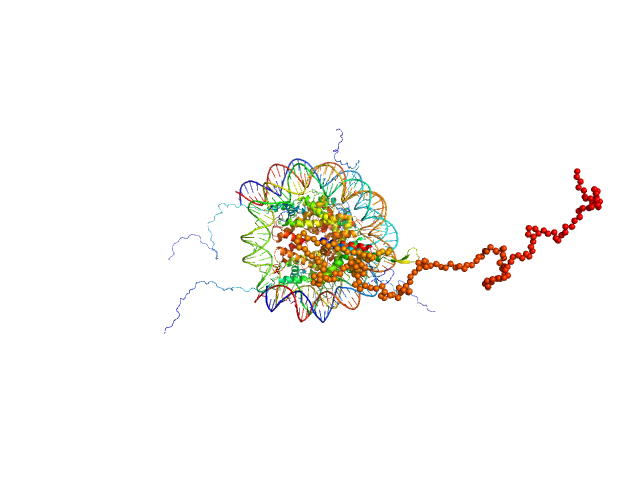
UniProt ID: Q4VC05-2 (1-231) Isoform 2 of B-cell CLL/lymphoma 7 protein family member A
UniProt ID: P84233 (1-135) Histone H3.2
UniProt ID: P62799 (1-103) Histone H4
UniProt ID: Q6AZJ8 (1-130) Histone H2A
UniProt ID: P02281 (1-126) Histone H2B 1.1
UniProt ID: None (None-None) Widom 601 DNA sequence
|
|
|
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.7 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
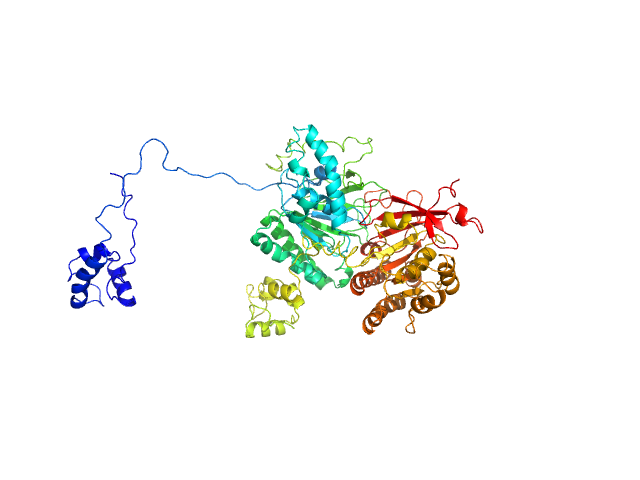
UniProt ID: O43502-1 (10-367) Isoform 1 of DNA repair protein RAD51 homolog 3
UniProt ID: O43542 (1-346) DNA repair protein XRCC3
|
|
|
|
| Sample: |
Isoform 1 of DNA repair protein RAD51 homolog 3 monomer, 40 kDa Homo sapiens protein
DNA repair protein XRCC3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 8, 100 mM NaCl, 2.5 mM ATP, 2.5 mM MgCl2, and 0.1 mM Na3VO4, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 May 5
|
RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles
Nature Communications 14(1) (2023)
Longo M, Roy S, Chen Y, Tomaszowski K, Arvai A, Pepper J, Boisvert R, Kunnimalaiyaan S, Keshvani C, Schild D, Bacolla A, Williams G, Tainer J, Schlacher K
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
UniProt ID: F5S9L7 (468-708) RTX toxin transporter
|
|
|
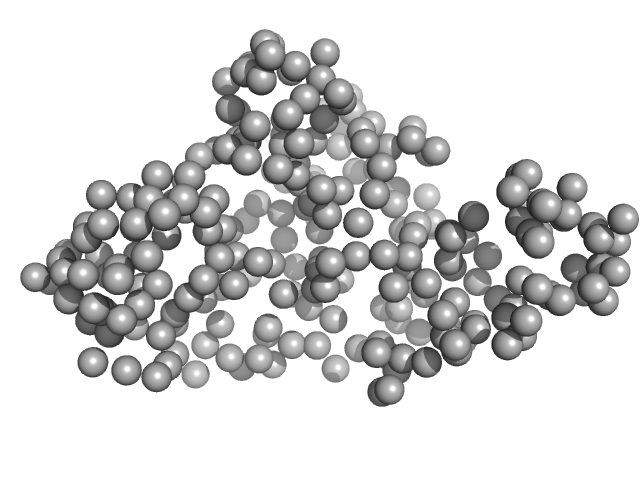
|
| Sample: |
RTX toxin transporter monomer, 28 kDa Kingella kingae ATCC … protein
|
| Buffer: |
100 mM HEPES, 10 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter
Scientific Reports 14(1) (2024)
Anlauf M, Bilsing F, Reiners J, Spitz O, Hachani E, Smits S, Schmitt L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: Q9KMA5 (2-104) Antitoxin HigA-2
|
|
|
|
| Sample: |
Antitoxin HigA-2 dimer, 23 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris pH 8, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Apr 6
|
Fuzzy recognition by the prokaryotic transcription factor HigA2 from Vibrio cholerae.
Nat Commun 15(1):3105 (2024)
Hadži S, Živič Z, Kovačič M, Zavrtanik U, Haeserts S, Charlier D, Plavec J, Volkov AN, Lah J, Loris R
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: Q9KMA5 (2-104) Antitoxin HigA-2
UniProt ID: None (None-None) DNA operator
|
|
|
|
| Sample: |
Antitoxin HigA-2 dimer, 23 kDa Vibrio cholerae serotype … protein
DNA operator dimer, 20 kDa DNA
|
| Buffer: |
20 mM Tris pH 8, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Apr 6
|
Fuzzy recognition by the prokaryotic transcription factor HigA2 from Vibrio cholerae.
Nat Commun 15(1):3105 (2024)
Hadži S, Živič Z, Kovačič M, Zavrtanik U, Haeserts S, Charlier D, Plavec J, Volkov AN, Lah J, Loris R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q9UMX0 (1-589) Ubiquilin-1
|
|
|
|
| Sample: |
Ubiquilin-1 dimer, 125 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 22
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Biophys J (2023)
Dao TP, Rajendran A, Galagedera SKK, Haws W, Castañeda CA
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
264 |
nm3 |
|
|