UniProt ID: Q8KC98 (None-None) Rab family protein
|
|
|

|
| Sample: |
Rab family protein dimer, 130 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Feb 1
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
UniProt ID: Q8KC98 (None-None) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 130 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µ M GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Feb 1
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
UniProt ID: Q8KC98 (None-None) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 130 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Feb 1
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
5.9 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
469 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
6.0 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
436 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
447 |
nm3 |
|
|
UniProt ID: P49419-2 (None-None) Aldehyde dehydrogenase 7A1 (Alpha-aminoadipic semialdehyde dehydrogenase)
|
|
|
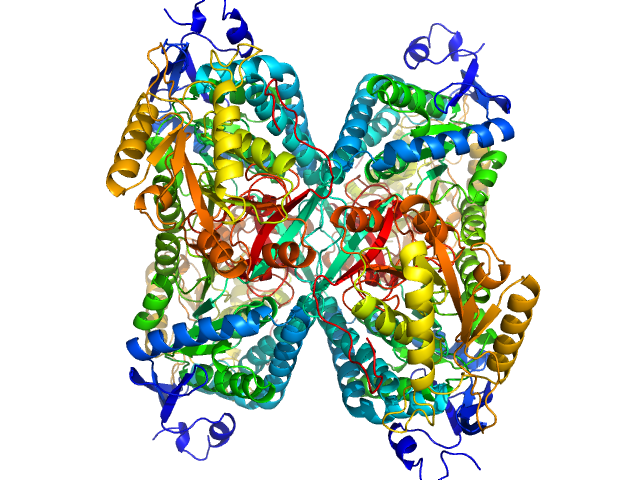
|
| Sample: |
Aldehyde dehydrogenase 7A1 (Alpha-aminoadipic semialdehyde dehydrogenase) tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 5% glycerol, 0.5 mM tris(3-hydroxypropyl)phosphine, 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 9
|
Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry 54(35):5513-22 (2015)
Luo M, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
UniProt ID: P04925 (23-230) Major prion protein
UniProt ID: (None-None) P-Clone Fab, Chimera
|
|
|
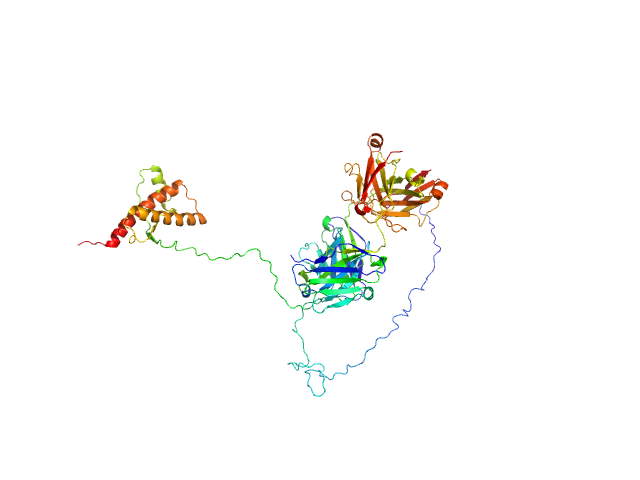
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
P-Clone Fab, Chimera monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
sodium acetate buffer (20 mM sodium acetate, pH 5.1; 150 mM NaCl), pH: 5.1 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2013 Dec 5
|
Prion Protein-Antibody Complexes Characterized by Chromatography-Coupled Small-Angle X-Ray Scattering.
Biophys J 109(4):793-805 (2015)
Carter L, Kim SJ, Schneidman-Duhovny D, Stöhr J, Poncet-Montange G, Weiss TM, Tsuruta H, Prusiner SB, Sali A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
UniProt ID: P24300 (1-388) Xylose isomerase
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
UniProt ID: Q9X721 (883-1118) Class1 collagenase
|
|
|

|
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|