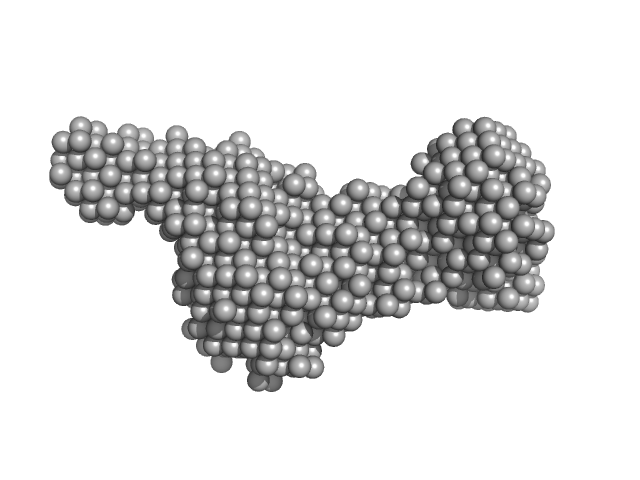
UniProt ID: Q9X721 (883-1118) Class1 collagenase collagen-binding domain
|
|
|
|
| Sample: |
Class1 collagenase collagen-binding domain monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: Q9X721 (883-1118) Class1 collagenase
|
|
|

|
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
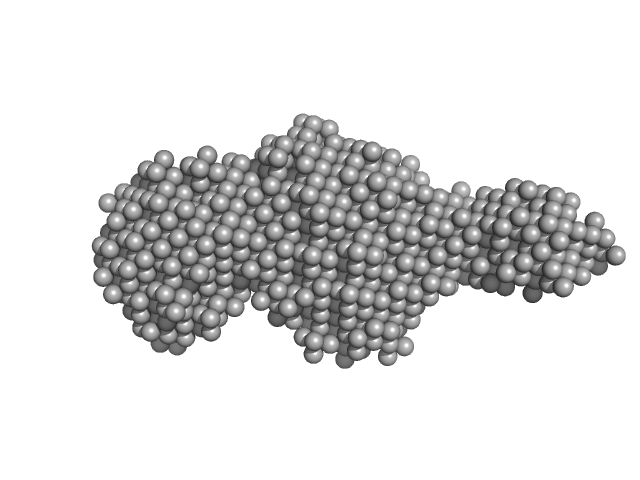
UniProt ID: Q9X721 (883-1118) Class1 collagenase
|
|
|
|
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P0DP33 (2-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Xenopus laevis protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
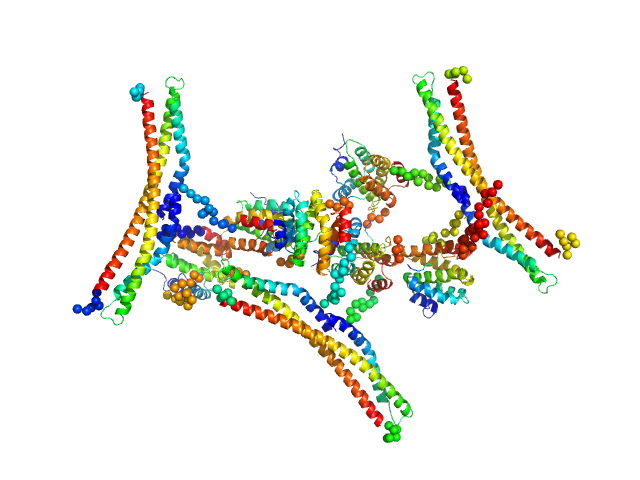
UniProt ID: Q05543 (1-246) Regulator of Ty1 transposition protein 103
UniProt ID: P04050 (None-None) DNA-directed RNA polymerase II subunit RPB1
|
|
|
|
| Sample: |
Regulator of Ty1 transposition protein 103 dimer, 57 kDa Saccharomyces cerevisiae protein
DNA-directed RNA polymerase II subunit RPB1 monomer, 13 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM KH2PO4, 300 mM KCl, 10 mM BME, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Apr 12
|
Structure and dynamics of the RNAPII CTDsome with Rtt103.
Proc Natl Acad Sci U S A 114(42):11133-11138 (2017)
Jasnovidova O, Klumpler T, Kubicek K, Kalynych S, Plevka P, Stefl R
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
561 |
nm3 |
|
|
UniProt ID: P08083 (1-387) Colicin N
|
|
|
|
| Sample: |
Colicin N monomer, 43 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: P08083 (40-387) Colicin N delta 1-39
|
|
|
|
| Sample: |
Colicin N delta 1-39 monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
UniProt ID: P08083 (1-387) Colicin N K145A mutant
|
|
|
|
| Sample: |
Colicin N K145A mutant monomer, 43 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: P08083 (1-90) Colicin N Translocation domain
|
|
|
|
| Sample: |
Colicin N Translocation domain monomer, 10 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: Q9J5G4 (1-143) Bcl-2-like protein FPV039
UniProt ID: Q5F404 (72-97) Uncharacterized protein (BAK1)
|
|
|
|
| Sample: |
Bcl-2-like protein FPV039 monomer, 17 kDa Fowlpox virus protein
Uncharacterized protein (BAK1) monomer, 3 kDa Gallus gallus protein
|
| Buffer: |
20 mM trisodium citrate pH, 200 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Oct 2
|
Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J Biol Chem 292(22):9010-9021 (2017)
Anasir MI, Caria S, Skinner MA, Kvansakul M
|
|
|