UniProt ID: P43320 (None-None) Beta-crystallin B2
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: P43320 (15-195) Beta-crystallin B2
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
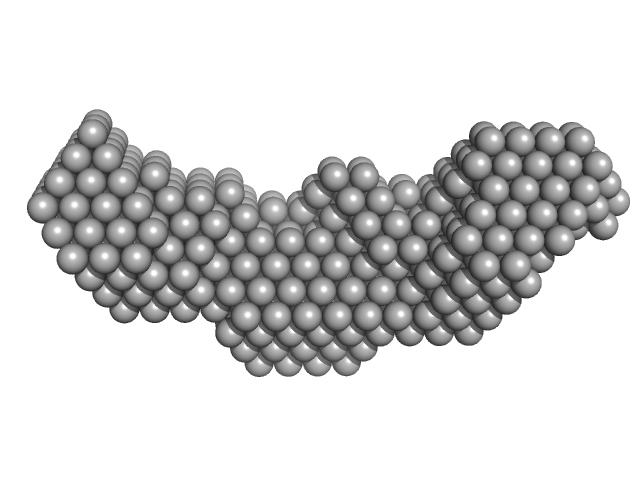
UniProt ID: P20271 (42-168) Homeobox protein CEH-14
UniProt ID: G5EC36 (347-376) CeLIM-7
|
|
|
|
| Sample: |
Homeobox protein CEH-14 monomer, 16 kDa Caenorhabditis elegans protein
CeLIM-7 monomer, 4 kDa Caenorhabditis elegans protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2009 Apr 7
|
Interactions between LHX3- and ISL1-family LIM-homeodomain transcription factors are conserved in Caenorhabditis elegans.
Sci Rep 7(1):4579 (2017)
Bhati M, Llamosas E, Jacques DA, Jeffries CM, Dastmalchi S, Ripin N, Nicholas HR, Matthews JM
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: P49333 (158-738) Ethylene Receptor 1
|
|
|
|
| Sample: |
Ethylene Receptor 1 dimer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris-NDSB 150 mM NaCl 1mM DTT 250 mM NDSB, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 19
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
UniProt ID: P49333 (158-738) Ethylene Receptor 1
|
|
|
|
| Sample: |
Ethylene Receptor 1 dimer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM TRIS 150 mM NaCl 1mM DTT 250mM NDSB, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 29
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
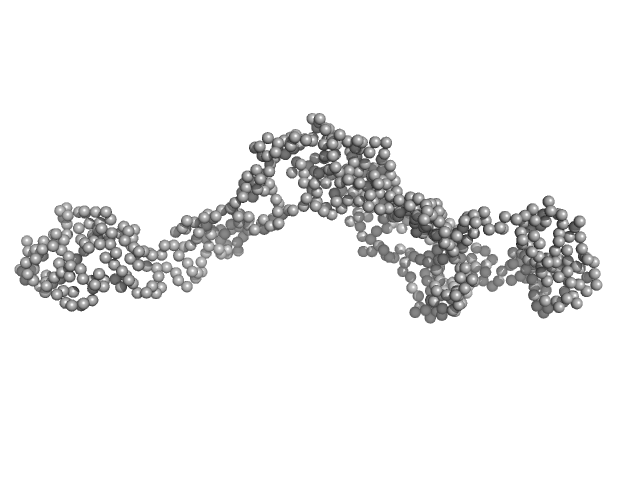
UniProt ID: Q06187 (None-None) Tyrosine-protein kinase BTK (R28C mutant)
|
|
|
|
| Sample: |
Tyrosine-protein kinase BTK (R28C mutant) monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, 2 mM DTT and 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Apr 2
|
Conformation of full-length Bruton tyrosine kinase (Btk) from synchrotron X-ray solution scattering.
EMBO J 22(18):4616-24 (2003)
Márquez JA, Smith CI, Petoukhov MV, Lo Surdo P, Mattsson PT, Knekt M, Westlund A, Scheffzek K, Saraste M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
UniProt ID: Q15554 (42-86) Basic domain of telomeric repeat-binding factor 2
|
|
|
|
| Sample: |
Basic domain of telomeric repeat-binding factor 2 monomer, 5 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaPi, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2015 Mar 5
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|

|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
6.2 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
354 |
nm3 |
|
|
UniProt ID: Q8KC98 (1-1102) Rab family protein
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
5.9 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
491 |
nm3 |
|
|