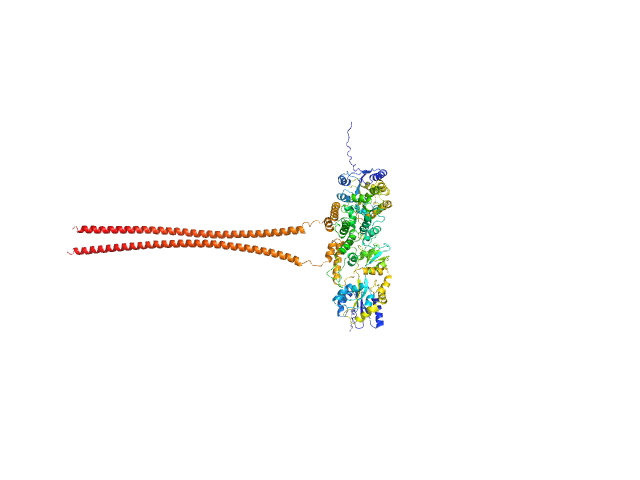
UniProt ID: G4VFI8 (307-412) Septin-10
|
|
|
|
| Sample: |
Septin-10 dimer, 110 kDa Schistosoma mansoni protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 17
|
Novel lipid-interaction motifs within the C-terminal domain of Septin10 from Schistosoma mansoni
Biochimica et Biophysica Acta (BBA) - Biomembranes :184371 (2024)
Cavini I, Fontes M, Zeraik A, Lopes J, Araújo A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
UniProt ID: None (None-None) Heterochromatin protein HP1α, S97A mutant, full-length
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Jun 2
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: None (None-None) phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Jun 2
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
3.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
UniProt ID: None (None-None) Heterochromatin protein HP1α, S97A mutant, full-length
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 500 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 19
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
UniProt ID: None (None-None) phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, S97A mutant, full-length dimer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 500 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 19
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
UniProt ID: None (1-113) Heterochromatin protein HP1α, delta CSD mutant
|
|
|
|
| Sample: |
Heterochromatin protein HP1α, delta CSD mutant monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 27
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
UniProt ID: None (1-113) phosphorylated Heterochromatin protein HP1α, delta CSD mutant
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, delta CSD mutant monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 27
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: None (1-113) phosphorylated Heterochromatin protein HP1α, delta CSD b4 mutant
|
|
|
|
| Sample: |
Phosphorylated Heterochromatin protein HP1α, delta CSD b4 mutant monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 27
|
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Nucleic Acids Research 53(6) (2025)
Furukawa A, Yonezawa K, Negami T, Yoshimura Y, Hayashi A, Nakayama J, Adachi N, Senda T, Shimizu K, Terada T, Shimizu N, Nishimura Y
|
| RgGuinier |
2.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
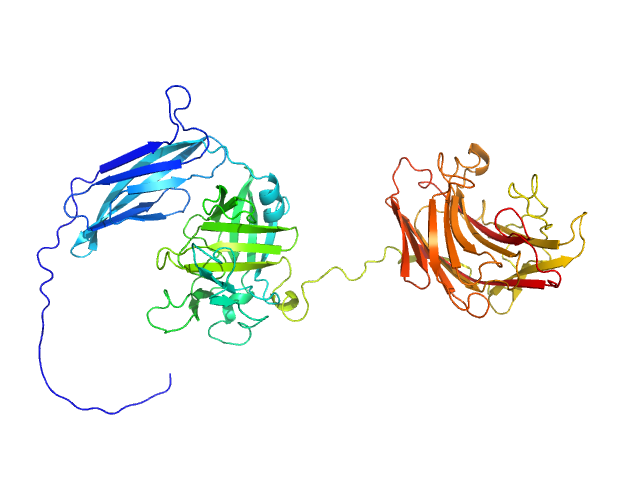
UniProt ID: A0M710-1 (29-538) Glycosyl hydrolase, family 16
|
|
|
|
| Sample: |
Glycosyl hydrolase, family 16 monomer, 58 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
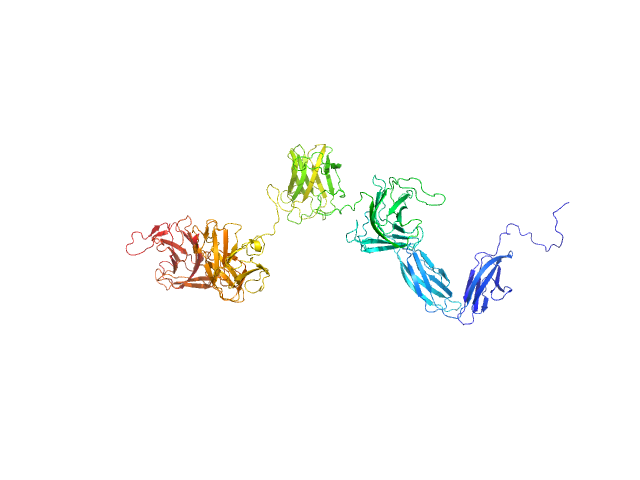
UniProt ID: A0M709 (21-896) PKD domain-containing protein
|
|
|
|
| Sample: |
PKD domain-containing protein monomer, 96 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
138 |
nm3 |
|
|