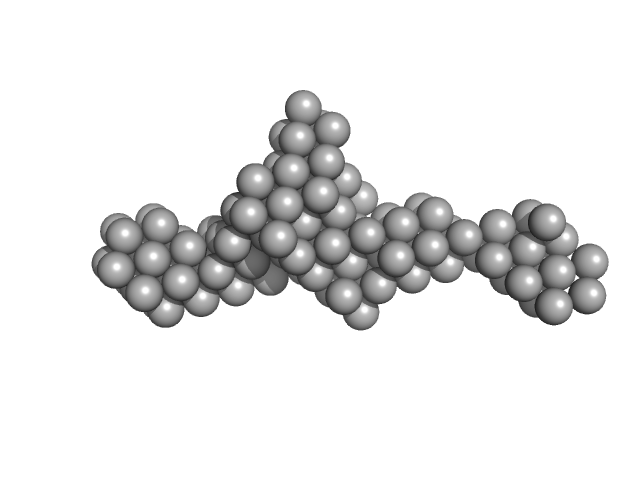
UniProt ID: Q9H171 (1-165) Z-DNA-binding protein 1
|
|
|
|
| Sample: |
Z-DNA-binding protein 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
ZBP1 condensate formation synergizes Z-NAs recognition and signal transduction.
Cell Death Dis 15(7):487 (2024)
Xie F, Wu D, Huang J, Liu X, Shen Y, Huang J, Su Z, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
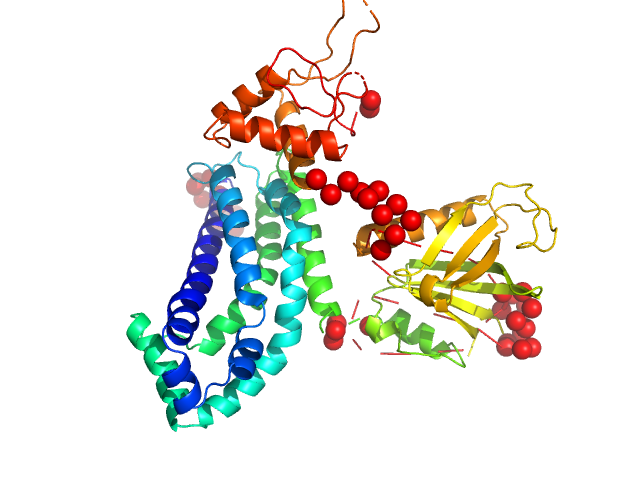
UniProt ID: Q8TCU6 (38-499) Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
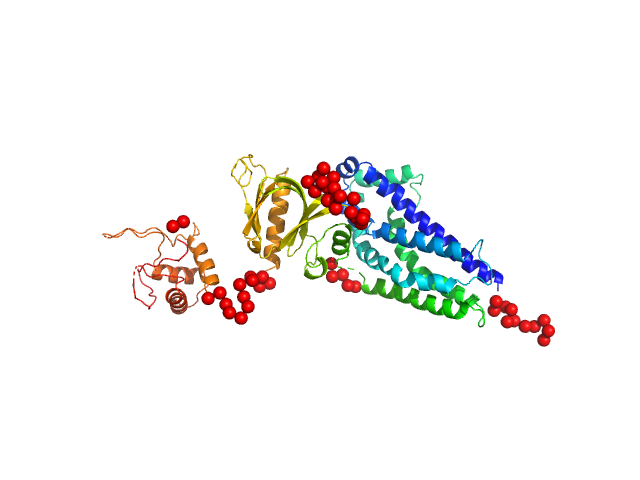
UniProt ID: Q8TCU6 (38-499) Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (A170K)
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (A170K) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 24
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
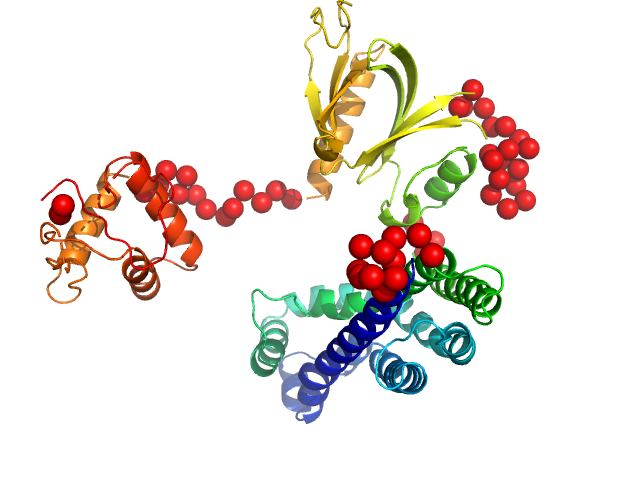
UniProt ID: Q8TCU6 (38-499) Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (L177E)
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (L177E) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
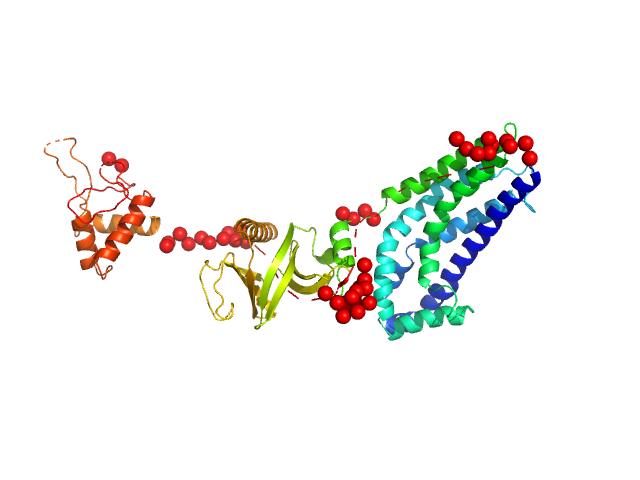
UniProt ID: Q8TCU6 (38-499) Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (I409A)
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (I409A) monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
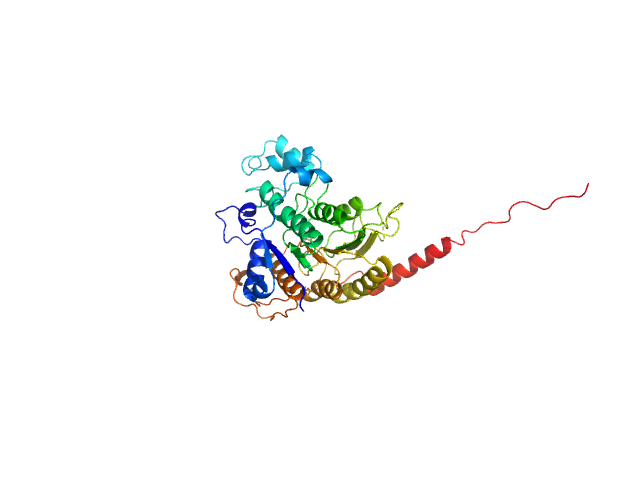
UniProt ID: Q9FH09 (5-383) Histone deacetylase 7
|
|
|
|
| Sample: |
Histone deacetylase 7 monomer, 44 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 24
|
Structure-function analyses reveal Arabidopsis thaliana HDA7 to be an inactive histone deacetylase
Current Research in Structural Biology :100136 (2024)
Saharan K, Baral S, Shaikh N, Vasudevan D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
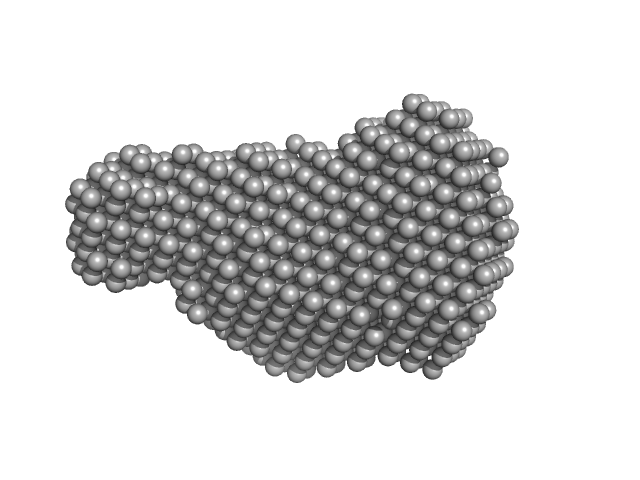
UniProt ID: Q8V4X6 (1-287) Virus termination factor small subunit
|
|
|
|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
UniProt ID: Q8V4X6 (1-287) Virus termination factor small subunit
UniProt ID: Q8V4Y7 (546-845) mRNA-capping enzyme catalytic subunit
|
|
|

|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
MRNA-capping enzyme catalytic subunit monomer, 35 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
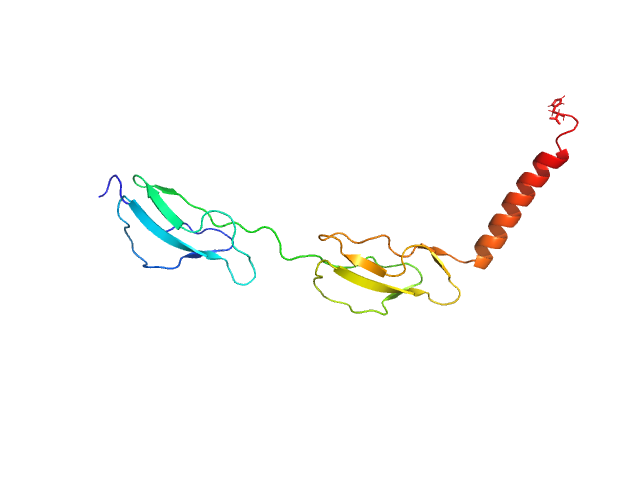
UniProt ID: P08603 (988-1107) Complement factor H
|
|
|
|
| Sample: |
Complement factor H monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 137 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 20
|
The SCR-17 and SCR-18 glycans in human complement Factor H enhance its regulatory function.
J Biol Chem :107624 (2024)
Gao X, Iqbal H, Yu DQ, Gor J, Coker AR, Perkins SJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
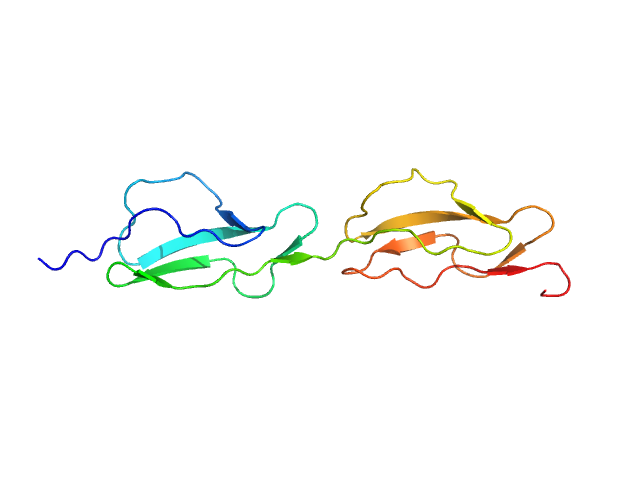
UniProt ID: P08603 (985-1108) Complement factor H
|
|
|
|
| Sample: |
Complement factor H monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 137 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 20
|
The SCR-17 and SCR-18 glycans in human complement Factor H enhance its regulatory function.
J Biol Chem :107624 (2024)
Gao X, Iqbal H, Yu DQ, Gor J, Coker AR, Perkins SJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|