UniProt ID: P0C1C6 (405-448) Protein W
|
|
|
|
| Sample: |
Protein W monomer, 6 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 16
|
Dissecting Henipavirus W proteins conformational and fibrillation properties: contribution of their N- and C-terminal constituent domains.
FEBS J (2024)
Pesce G, Gondelaud F, Ptchelkine D, Bignon C, Fourquet P, Longhi S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
UniProt ID: P0C1C7 (407-450) Protein W
|
|
|
|
| Sample: |
Protein W monomer, 6 kDa Henipavirus nipahense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 16
|
Dissecting Henipavirus W proteins conformational and fibrillation properties: contribution of their N- and C-terminal constituent domains.
FEBS J (2024)
Pesce G, Gondelaud F, Ptchelkine D, Bignon C, Fourquet P, Longhi S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
UniProt ID: A0A349B205 (1-608) GGDEF domain-containing protein
|
|
|
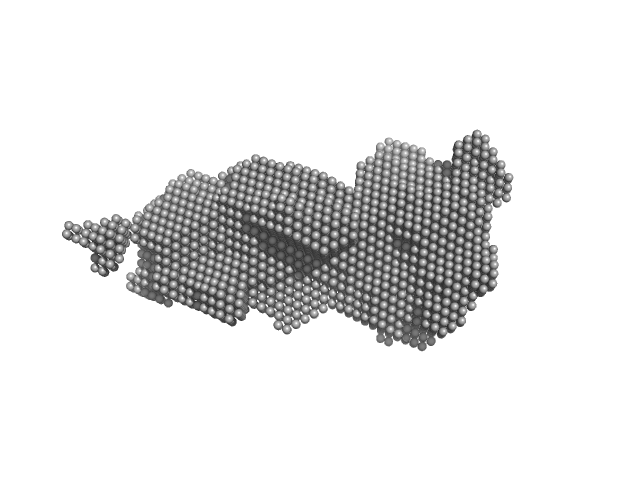
|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
UniProt ID: A0A349B205 (1-608) GGDEF domain-containing protein
|
|
|

|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
UniProt ID: E7FRT5 (35-472) LPXTG-motif cell wall anchor domain protein
|
|
|
|
| Sample: |
LPXTG-motif cell wall anchor domain protein monomer, 49 kDa Ligilactobacillus ruminis ATCC … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 1
|
The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis
Acta Crystallographica Section D Structural Biology 80(7):474-492 (2024)
Prajapati A, Palva A, von Ossowski I, Krishnan V
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
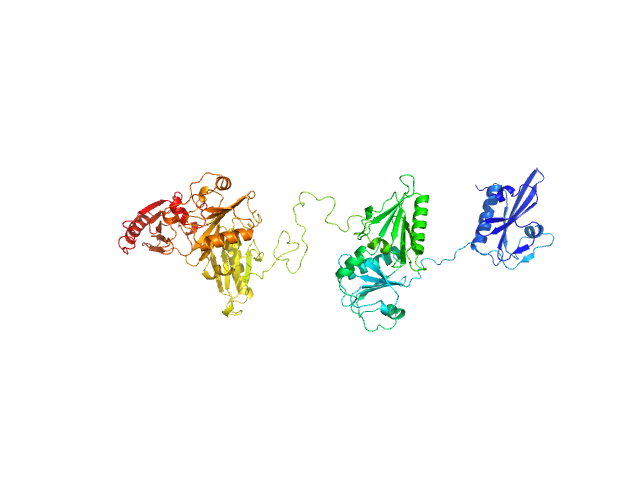
UniProt ID: P06396 (25-782) Gelsolin
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, pH8, 45 mM NaCl, 1 mM EGTA + 4 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|

|
| Sample: |
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
13.4 |
nm |
| Dmax |
48.0 |
nm |
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|
UniProt ID: P06396 (25-782) Gelsolin
UniProt ID: P60706 (1-375) Actin, cytoplasmic 1
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 18-mer, 751 kDa Gallus gallus protein
|
| Buffer: |
50 mM KCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM ATP, 0.1% 2-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
|
|