UniProt ID: O65258 (56-542) Beta-amylase 2, chloroplastic
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
UniProt ID: O65258 (56-542) Beta-amylase 2, chloroplastic
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
UniProt ID: O65258 (56-542) Beta-amylase 2, chloroplastic
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
UniProt ID: O65258 (56-542) Beta-amylase 2, chloroplastic
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
UniProt ID: Q9P8F4 (1-860) beta-glucosidase
|
|
|
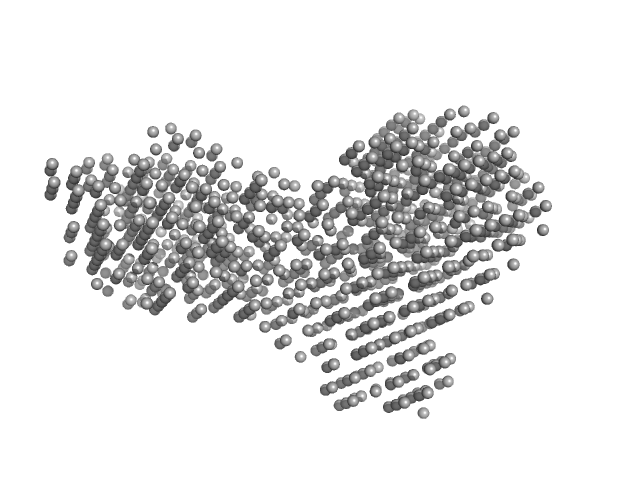
|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jun 4
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
UniProt ID: P69905 (2-142) Hemoglobin subunit alpha
UniProt ID: P68871 (2-147) Hemoglobin subunit beta
UniProt ID: None (None-None) Protoporphyrin IX containing fe
|
|
|
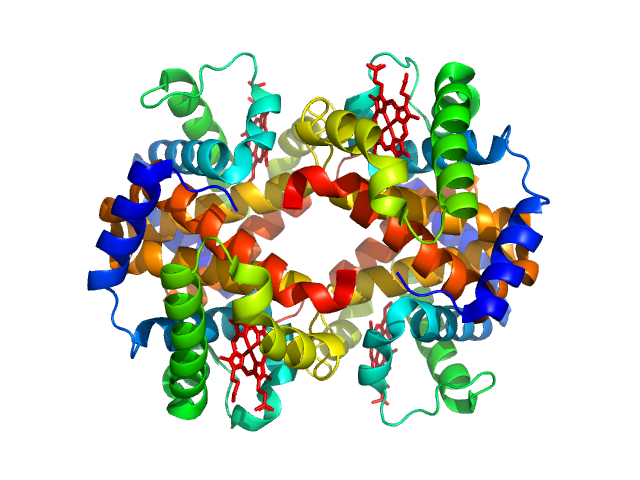
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100 mM Sodium Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 May 24
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
UniProt ID: P69905 (2-142) Hemoglobin subunit alpha
UniProt ID: P68871 (2-147) Hemoglobin subunit beta
UniProt ID: None (None-None) Protoporphyrin IX containing fe
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100 mM Sodium Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Sep 30
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: P69905 (2-142) Hemoglobin subunit alpha
UniProt ID: P68871 (2-147) Hemoglobin subunit beta
UniProt ID: None (None-None) Protoporphyrin IX containing fe
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 5% (w/v) PEG600, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Sep 30
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
UniProt ID: P69905 (2-142) Hemoglobin subunit alpha
UniProt ID: P68871 (2-147) Hemoglobin subunit beta
UniProt ID: None (None-None) Protoporphyrin IX containing fe
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 10% (w/v) PEG600, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Sep 30
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
UniProt ID: P69905 (2-142) Hemoglobin subunit alpha
UniProt ID: P68871 (2-147) Hemoglobin subunit beta
UniProt ID: None (None-None) Protoporphyrin IX containing fe
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 20% (w/v) PEG600, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Sep 30
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
113 |
nm3 |
|
|