UniProt ID: P0AEX9 (27-384) Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A)
UniProt ID: P9WPH5 (19-409) Uncharacterized protein Rv2242
|
|
|
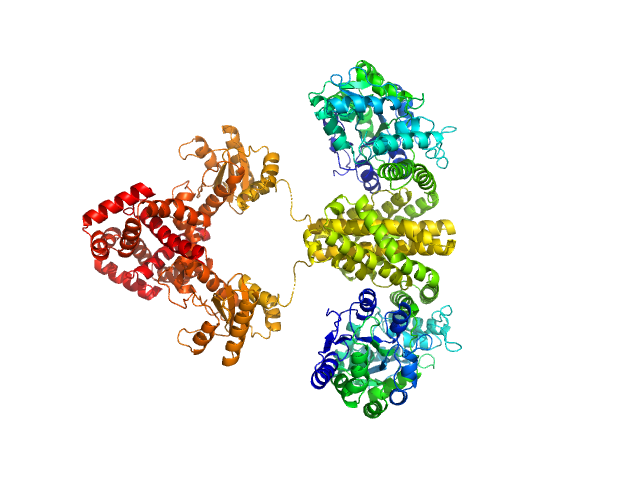
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) dimer, 80 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 dimer, 86 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
345 |
nm3 |
|
|
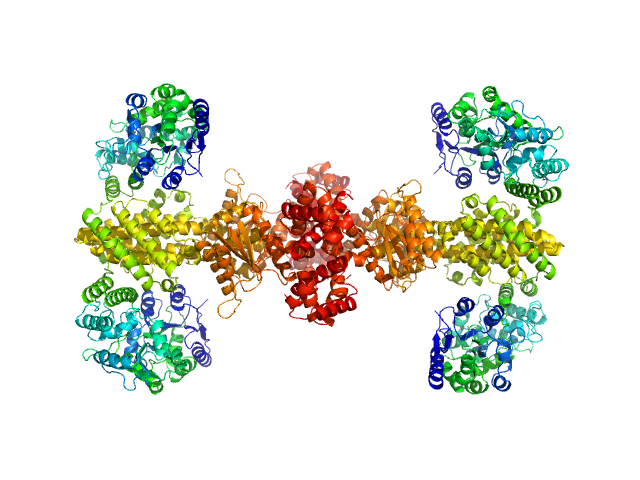
UniProt ID: P0AEX9 (27-384) Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A)
UniProt ID: P9WPH5 (19-409) Uncharacterized protein Rv2242
|
|
|
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) tetramer, 161 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 tetramer, 171 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
826 |
nm3 |
|
|
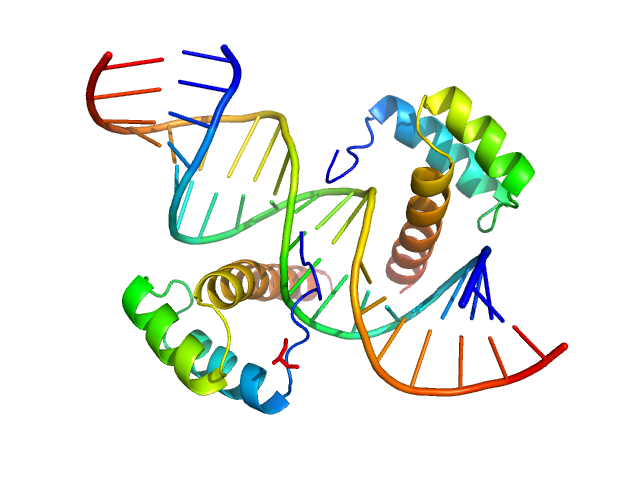
UniProt ID: O43186 (31-107) Cone-rod homeobox protein
UniProt ID: None (None-None) Ret4 sense strand
UniProt ID: None (None-None) Ret4 antisense strand
|
|
|
|
| Sample: |
Cone-rod homeobox protein, 20 kDa Homo sapiens protein
Ret4 sense strand, 6 kDa synthetic construct DNA
Ret4 antisense strand, 6 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 25
|
Molecular basis of CRX/DNA recognition and stoichiometry at the Ret4 response element
Structure (2024)
Srivastava D, Gowribidanur-Chinnaswamy P, Gaur P, Spies M, Swaroop A, Artemyev N
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: P40818 (7-1110) Ubiquitin carboxyl-terminal hydrolase 8
|
|
|

|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 8 monomer, 127 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 18
|
Autoinhibition of ubiquitin-specific protease 8: insights into domain interactions and mechanisms of regulation
Journal of Biological Chemistry :107727 (2024)
Caba C, Black M, Liu Y, DaDalt A, Mallare J, Fan L, Harding R, Wang Y, Vacratsis P, Huang R, Zhuang Z, Tong Y
|
| RgGuinier |
8.4 |
nm |
| Dmax |
31.8 |
nm |
| VolumePorod |
327 |
nm3 |
|
|
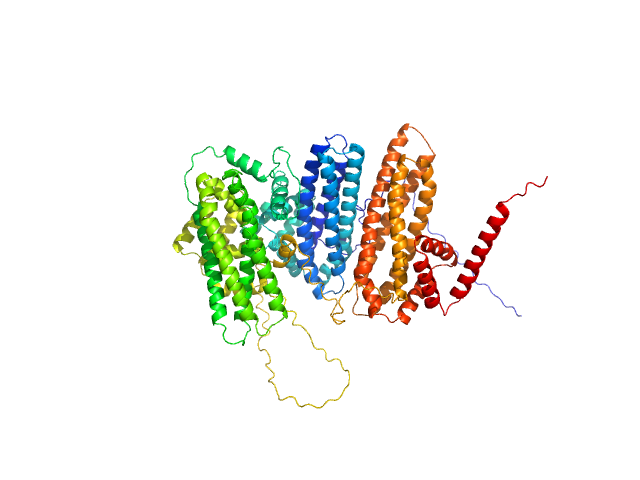
UniProt ID: F4IDY5 (1-831) Zinc finger protein BRUTUS-like At1g18910
|
|
|
|
| Sample: |
Zinc finger protein BRUTUS-like At1g18910 monomer, 98 kDa Arabidopsis thaliana protein
|
| Buffer: |
10 mM MES, 15 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 8
|
Iron-sensing and redox properties of the hemerythrin-like domains of Arabidopsis BRUTUS and BRUTUS-LIKE2 proteins.
Nat Commun 16(1):3865 (2025)
Pullin J, Rodríguez-Celma J, Franceschetti M, Mundy JEA, Svistunenko DA, Bradley JM, Le Brun NE, Balk J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
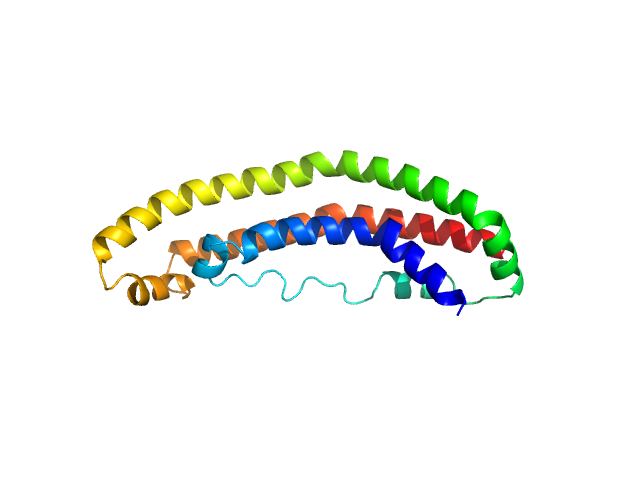
UniProt ID: P02647 (25-208) Apolipoprotein A-I
|
|
|
|
| Sample: |
Apolipoprotein A-I monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 20
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P02647 (25-208) Apolipoprotein A-I
|
|
|

|
| Sample: |
Apolipoprotein A-I dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 20
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: P02647 (25-208) Apolipoprotein A-I
|
|
|
|
| Sample: |
Apolipoprotein A-I monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 1
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P02647 (25-208) Apolipoprotein A-I (G50R)
|
|
|
|
| Sample: |
Apolipoprotein A-I (G50R) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Jun 27
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
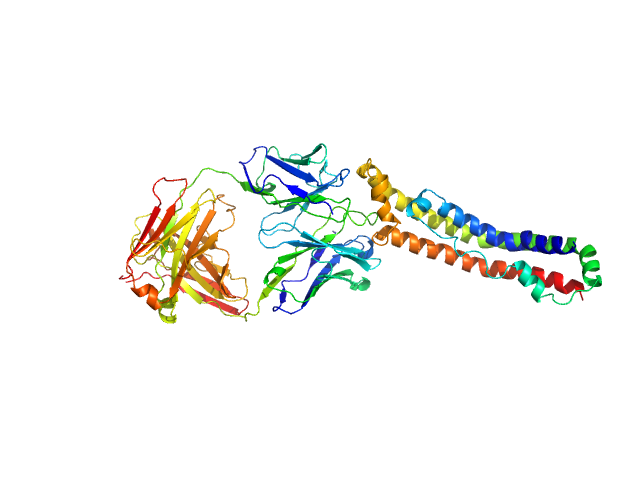
UniProt ID: P02647 (25-267) Apolipoprotein A-I
UniProt ID: None (None-None) Antigen-binding fragment 55201
|
|
|
|
| Sample: |
Apolipoprotein A-I monomer, 28 kDa Homo sapiens protein
Antigen-binding fragment 55201 monomer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 19
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
99 |
nm3 |
|
|