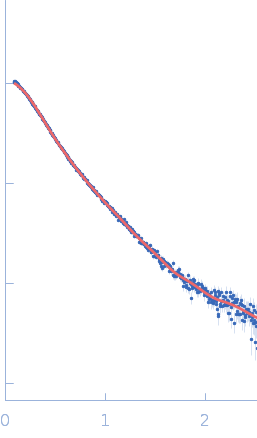
UniProt ID: Q9Y371 (1-306) Endophilin-B1 (Δ307-360)
|
|
|
|
| Sample: |
Endophilin-B1 (Δ307-360) dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
UniProt ID: O49003 (404-546) non-specific serine/threonine protein kinase
|
|
|
|
| Sample: |
Non-specific serine/threonine protein kinase monomer, 17 kDa Avena sativa protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2021 May 15
|
Structural dynamics of protein-protein association involved in the light-induced transition of Avena sativa LOV2 protein.
Nat Commun 15(1):6991 (2024)
Kim C, Yun SR, Lee SJ, Kim SO, Lee H, Choi J, Kim JG, Kim TW, You S, Kosheleva I, Noh T, Baek J, Ihee H
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: O49003 (404-546) non-specific serine/threonine protein kinase
|
|
|
|
| Sample: |
Non-specific serine/threonine protein kinase dimer, 33 kDa Avena sativa protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2021 May 15
|
Structural dynamics of protein-protein association involved in the light-induced transition of Avena sativa LOV2 protein.
Nat Commun 15(1):6991 (2024)
Kim C, Yun SR, Lee SJ, Kim SO, Lee H, Choi J, Kim JG, Kim TW, You S, Kosheleva I, Noh T, Baek J, Ihee H
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
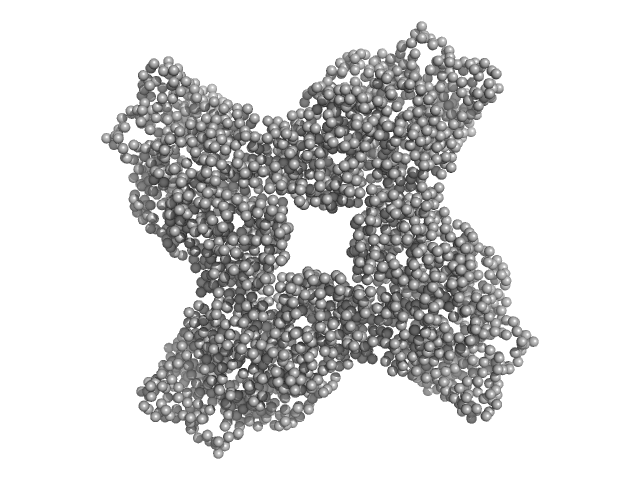
UniProt ID: E0U0N4 (2-657) Putative acylaminoacyl-peptidase
|
|
|
|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
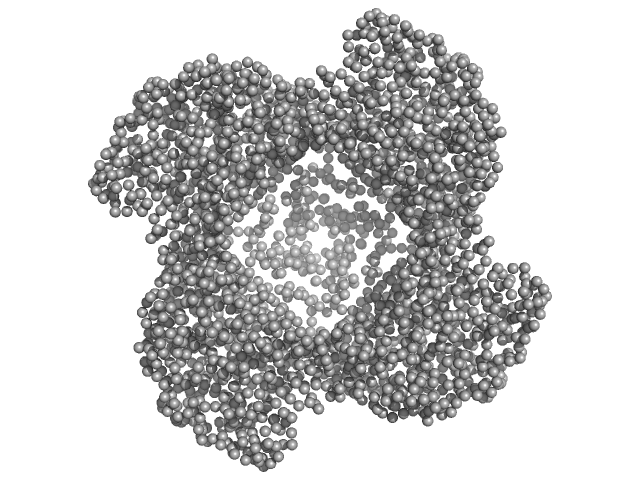
UniProt ID: E0U0N4 (2-657) Putative acylaminoacyl-peptidase
|
|
|
|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
435 |
nm3 |
|
|
UniProt ID: E0U0N4 (2-657) Putative acylaminoacyl-peptidase
|
|
|

|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
UniProt ID: P03372 (1-184) Estrogen receptor
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jul 17
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
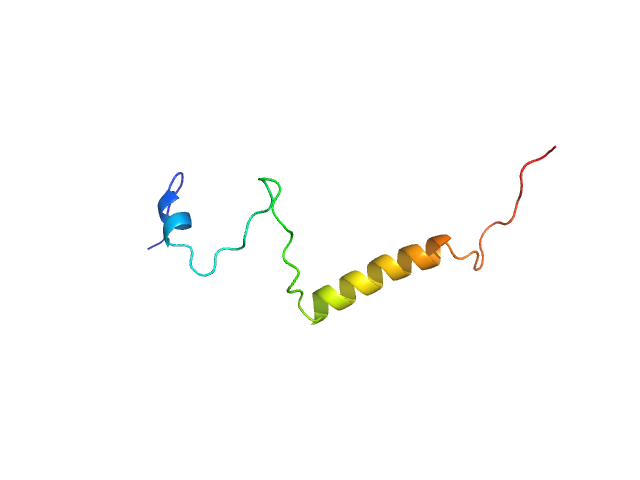
UniProt ID: O75920-2 (1-62) Isoform Short of Small EDRK-rich factor 1
|
|
|
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Mar 11
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
UniProt ID: O75920-2 (1-62) Isoform Short of Small EDRK-rich factor 1
UniProt ID: None (None-None) NT17
|
|
|
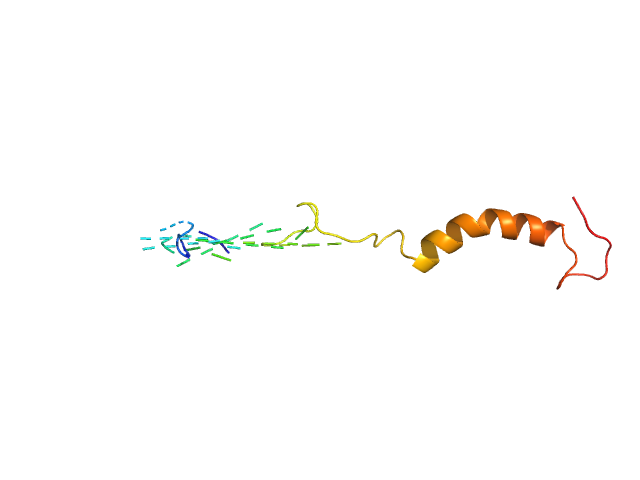
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
NT17 dimer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
|
|
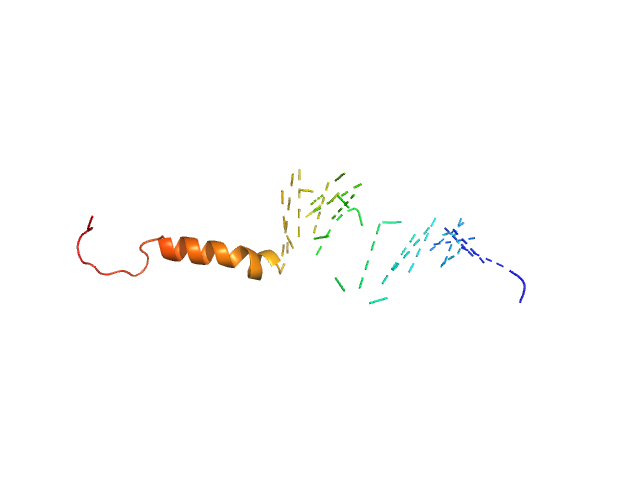
UniProt ID: O75920-2 (1-62) Isoform Short of Small EDRK-rich factor 1
UniProt ID: None (None-None) HTT3
|
|
|
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
HTT3 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 May 20
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
9 |
nm3 |
|
|