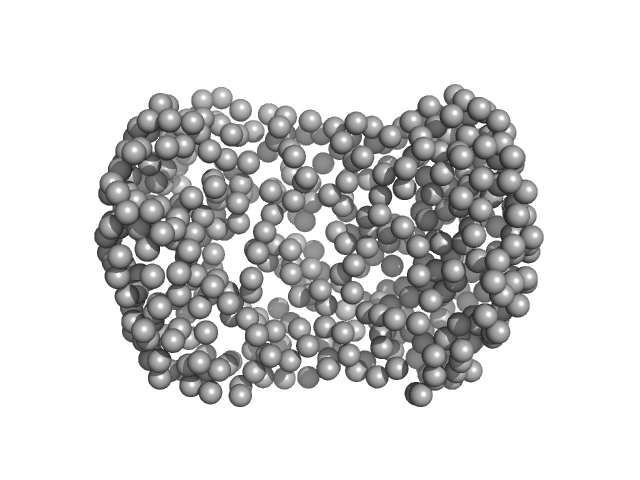
UniProt ID: A0A0G3FJH3 (52-470) Auxin response factor
|
|
|
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
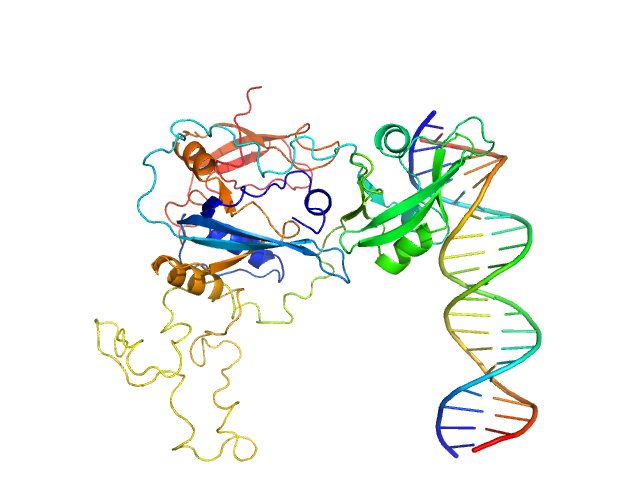
UniProt ID: A0A0G3FJH3 (52-470) Auxin response factor
UniProt ID: None (None-None) High Affinity ARF binding sequence inverted repeat with 6 nucleotide spacing
|
|
|
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
High Affinity ARF binding sequence inverted repeat with 6 nucleotide spacing dimer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: Q8L7G0 (1-355) Auxin response factor 1
UniProt ID: None (None-None) High Affinity ARF binding sequence inverted repeat with 7 nucleotide spacing
|
|
|
|
| Sample: |
Auxin response factor 1 dimer, 81 kDa Arabidopsis thaliana protein
High Affinity ARF binding sequence inverted repeat with 7 nucleotide spacing dimer, 13 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
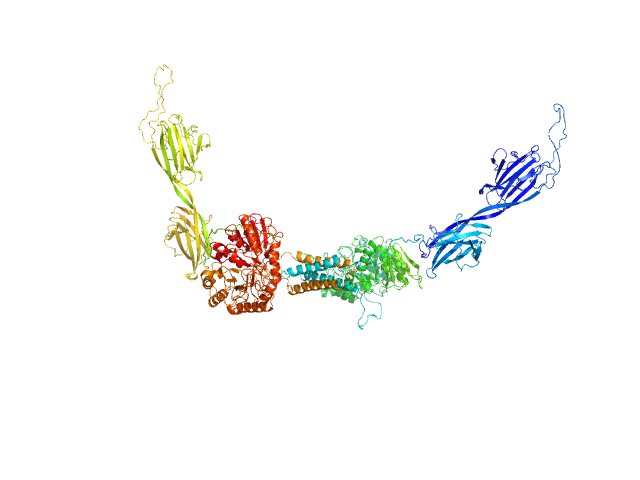
UniProt ID: Q94A41 (57-887) Alpha-amylase 3, chloroplastic
|
|
|
|
| Sample: |
Alpha-amylase 3, chloroplastic dimer, 187 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
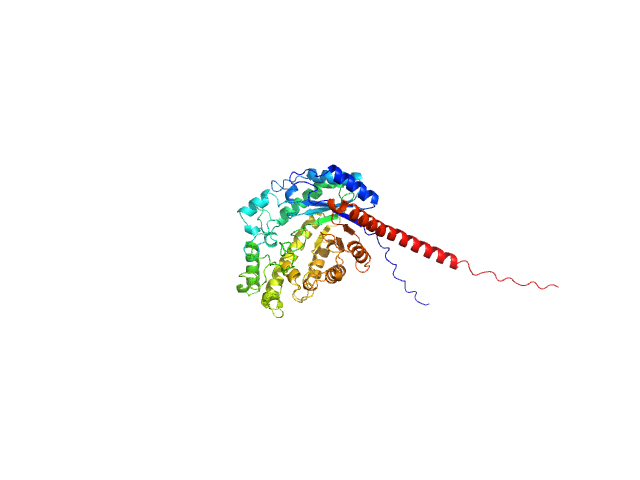
UniProt ID: Q8VYW2 (73-536) Inactive beta-amylase 9
|
|
|
|
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: Q8VYW2 (73-536) Inactive beta-amylase 9
UniProt ID: Q94A41 (57-887) Alpha-amylase 3, chloroplastic
|
|
|
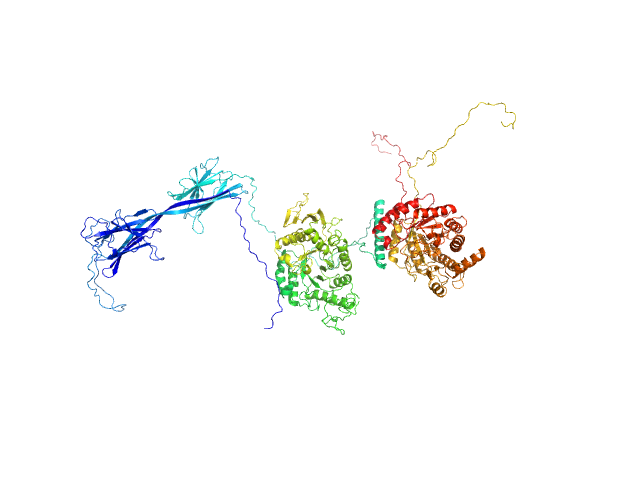
|
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
Alpha-amylase 3, chloroplastic monomer, 94 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
UniProt ID: P43351 (1-212) DNA repair protein RAD52 homolog
|
|
|

|
| Sample: |
DNA repair protein RAD52 homolog decamer, 234 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-Tris, 10% glycerol, 400 mM NaCl, 100 mM KCl, 1 mM EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS 1000, Eppley Structural Biology Facility, University of Nebraska Medical Center on 2014 Jul 25
|
A glimpse into the hidden world of the flexible C-terminal protein binding domains of human RAD52
Journal of Structural Biology 216(3):108115 (2024)
Struble L, Lovelace J, Borgstahl G
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
375 |
nm3 |
|
|
UniProt ID: P43351 (1-303) DNA repair protein RAD52 homolog
|
|
|

|
| Sample: |
DNA repair protein RAD52 homolog nonamer, 300 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-Tris, 10% GLycerol, 400 mM NaCl, 100 mM KCl, 1mM EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS 1000, Eppley Structural Biology Facility, University of Nebraska Medical Center on 2014 Jul 25
|
A glimpse into the hidden world of the flexible C-terminal protein binding domains of human RAD52
Journal of Structural Biology 216(3):108115 (2024)
Struble L, Lovelace J, Borgstahl G
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
782 |
nm3 |
|
|
UniProt ID: P10636-8 (244-372) Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A)
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P10636-8 (244-372) Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A)
UniProt ID: A0A091E260 (1-77) Polyubiquitin-B
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|