UniProt ID: Q8XAD7 (1-100) Phage antirepressor protein Cro
UniProt ID: None (None-None) IR1 22 base pair dsDNA
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDV54_fit1_model1.png)
|
| Sample: |
Phage antirepressor protein Cro dimer, 24 kDa Escherichia coli O157:H7 protein
IR1 22 base pair dsDNA monomer, 14 kDa DNA
|
| Buffer: |
20 mM sodium acetate, 150 mM NaCl, 1 mM TCEP, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 13
|
Structural-function relationship of YdaS, a Cro-type repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7
Marusa Prolic Kalinsek
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: A0A0D6HZA6 (1-246) Alpha/beta fold hydrolase
|
|
|

|
| Sample: |
Alpha/beta fold hydrolase monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM NaCl, 10 mM HEPES, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Apr 11
|
Similar but Distinct—Biochemical Characterization of the
Staphylococcus aureus
Serine Hydrolases FphH
and FphI
Proteins: Structure, Function, and Bioinformatics (2024)
Fellner M, Randall G, Bitac I, Warrender A, Sethi A, Jelinek R, Kass I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: Q13148 (1-80) TAR DNA-binding protein 43
|
|
|
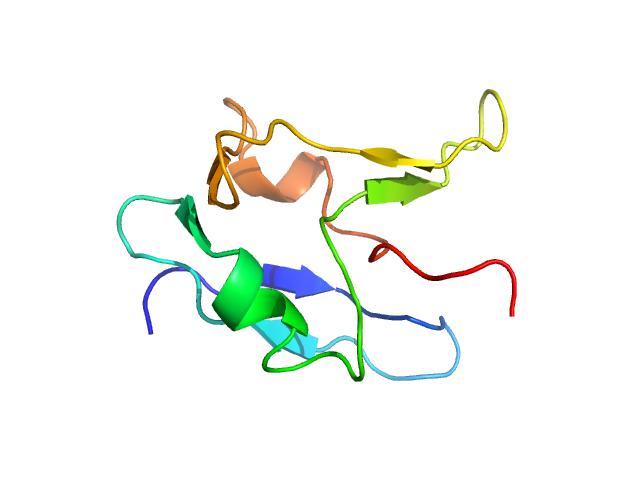
|
| Sample: |
TAR DNA-binding protein 43 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2024 May 29
|
Semi-synthesis of TDP-43 reveals the effects of phosphorylation in N-terminal domain on self-association
Communications Chemistry 8(1) (2025)
Sasaki D, Tenda M, Sohma Y
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
UniProt ID: Q9Y3A5 (1-250) Ribosome maturation protein SBDS
|
|
|
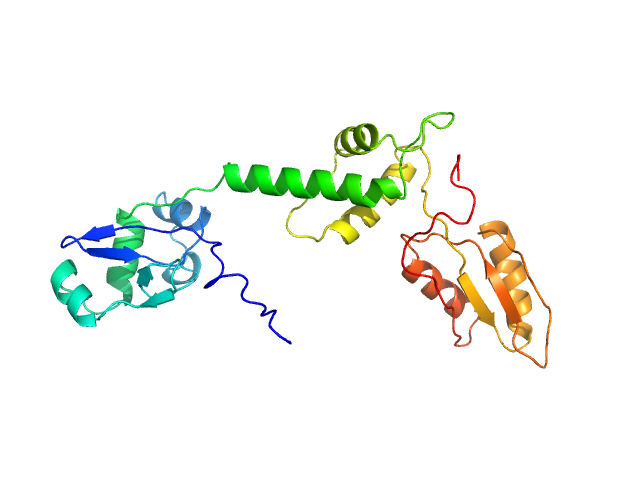
|
| Sample: |
Ribosome maturation protein SBDS monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: Q9Y3A5 (1-250) Ribosome maturation protein SBDS (R19Q)
|
|
|
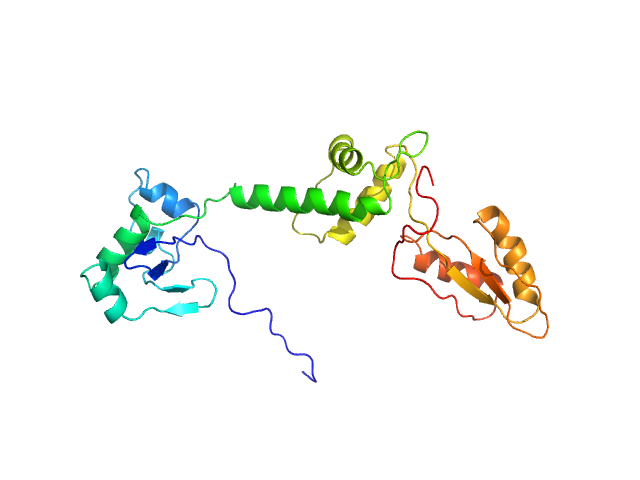
|
| Sample: |
Ribosome maturation protein SBDS (R19Q) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: Q9Y3A5 (1-250) Ribosome maturation protein SBDS (I167T)
|
|
|
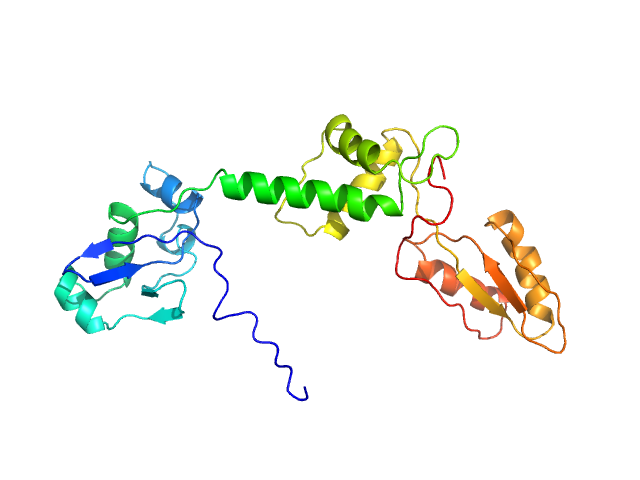
|
| Sample: |
Ribosome maturation protein SBDS (I167T) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
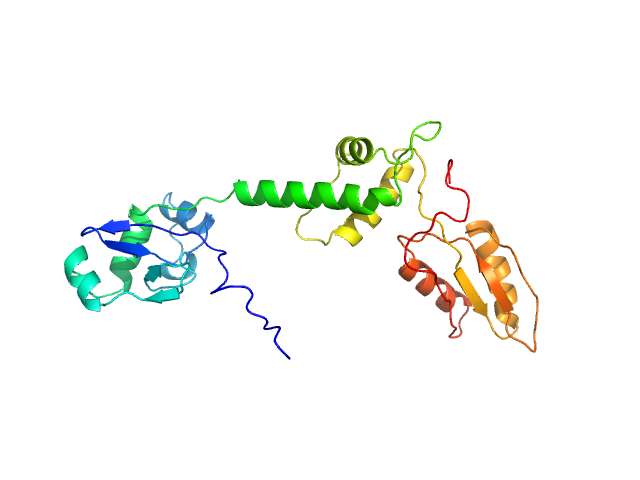
UniProt ID: Q9Y3A5 (1-250) Ribosome maturation protein SBDS (R175W)
|
|
|
|
| Sample: |
Ribosome maturation protein SBDS (R175W) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
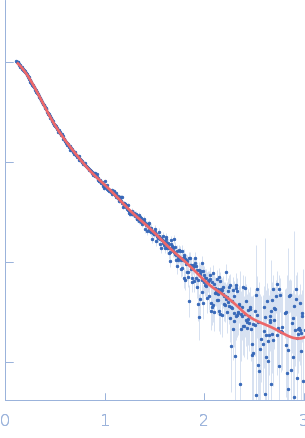
UniProt ID: Q9Y371 (1-365) Endophilin-B1
|
|
|
|
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
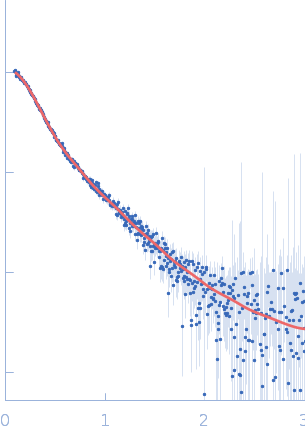
UniProt ID: Q9Y371 (1-365) Endophilin-B1
|
|
|
|
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
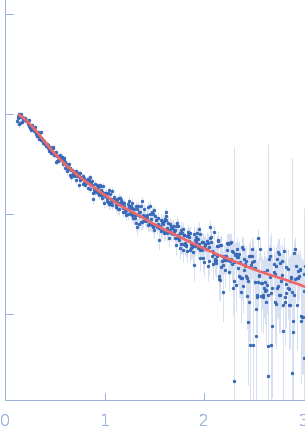
UniProt ID: Q9Y371 (1-365) Endophilin-B1
|
|
|
|
| Sample: |
Endophilin-B1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|