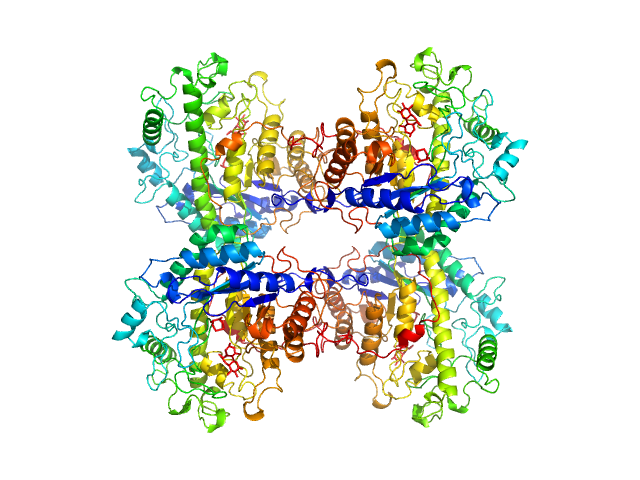
UniProt ID: P10537 (1-499) Beta-amylase
|
|
|
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
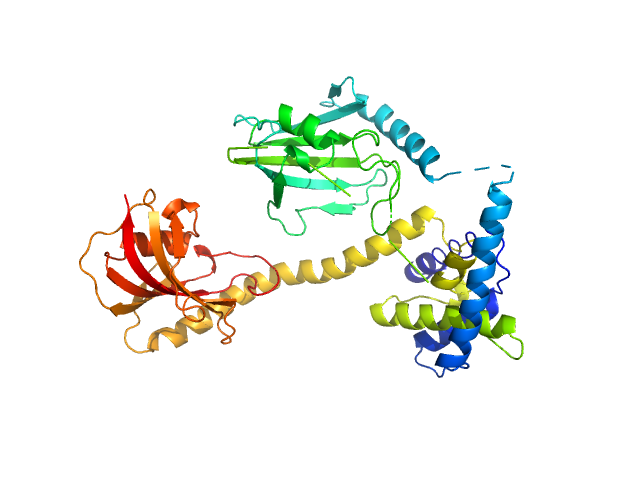
UniProt ID: Q5ZXE0 (21-233) Outer membrane protein MIP
UniProt ID: None (None-None) (2S)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐4‐methyl‐N‐[(pyridin‐3‐ yl)methyl]pentanamide
|
|
|
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐4‐methyl‐N‐[(pyridin‐3‐ yl)methyl]pentanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
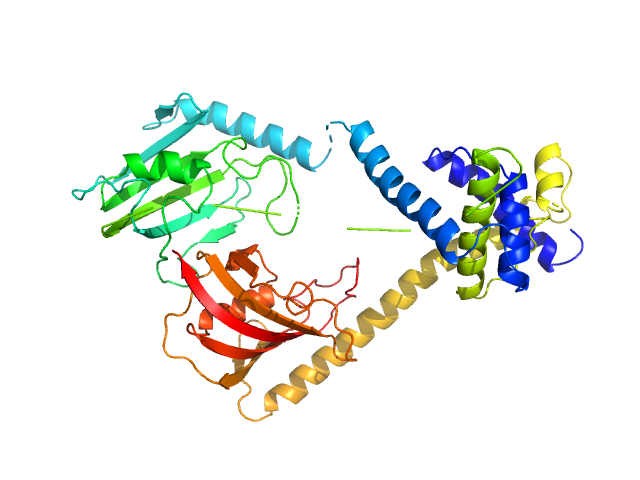
UniProt ID: Q5ZXE0 (21-233) Outer membrane protein MIP
UniProt ID: None (None-None) (2S)‐3‐(4‐fluorophenyl)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐N‐[(pyridin‐3‐yl)methyl]propanamide
|
|
|
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐3‐(4‐fluorophenyl)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐N‐[(pyridin‐3‐yl)methyl]propanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
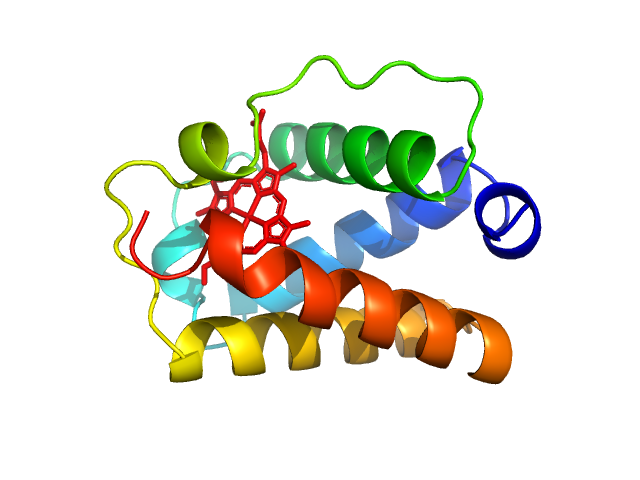
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 May 1
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|