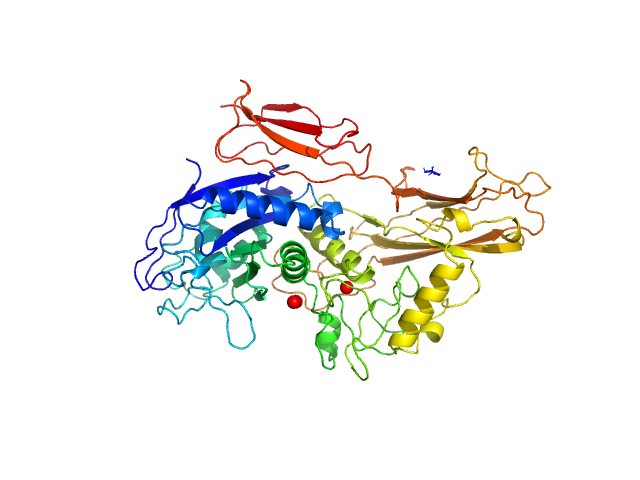
UniProt ID: Q9AES2 (329-876) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 63 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
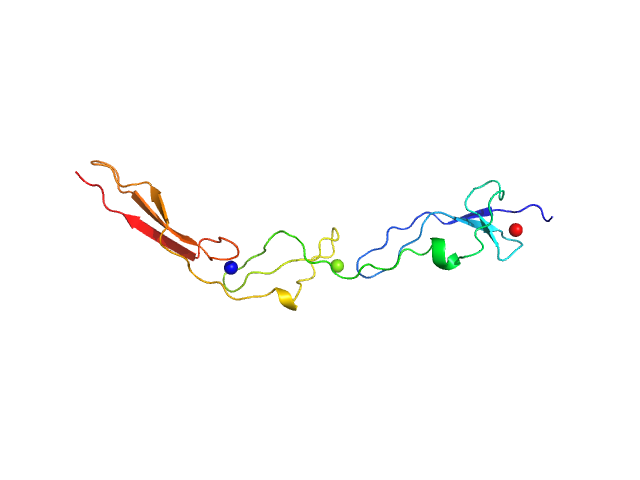
UniProt ID: Q9AES2 (880-1008) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 14 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
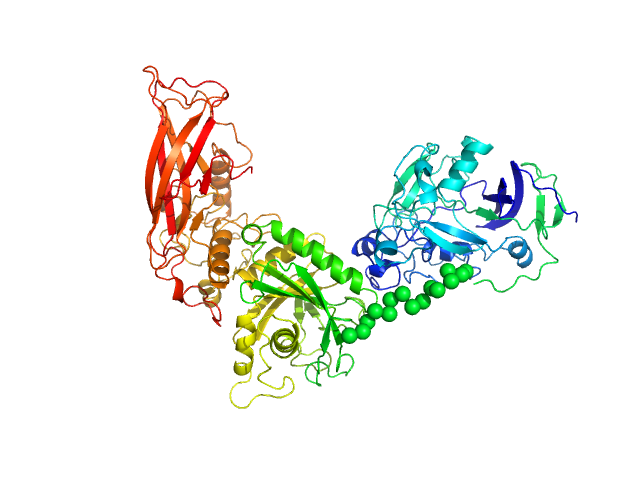
UniProt ID: Q9AES2 (31-807) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 88 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
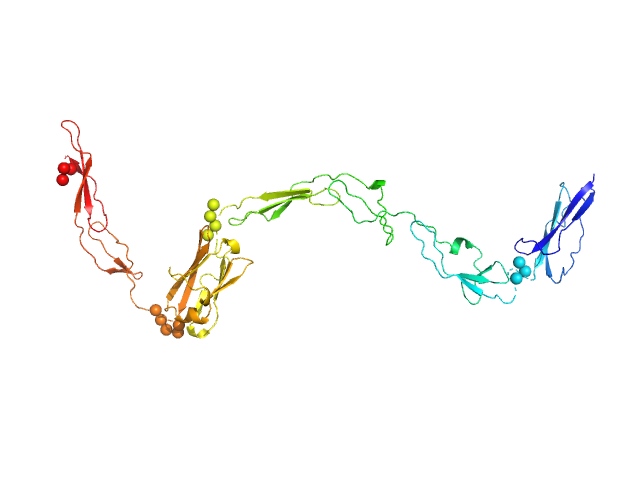
UniProt ID: Q9AES2 (808-1170) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 39 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
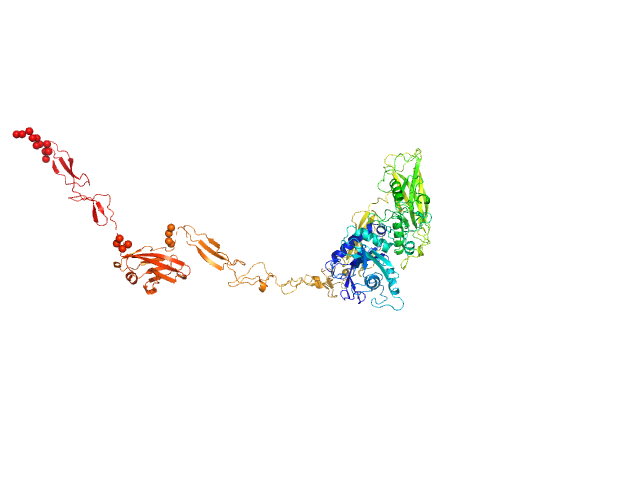
UniProt ID: Q9AES2 (329-1170) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 95 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
5.7 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
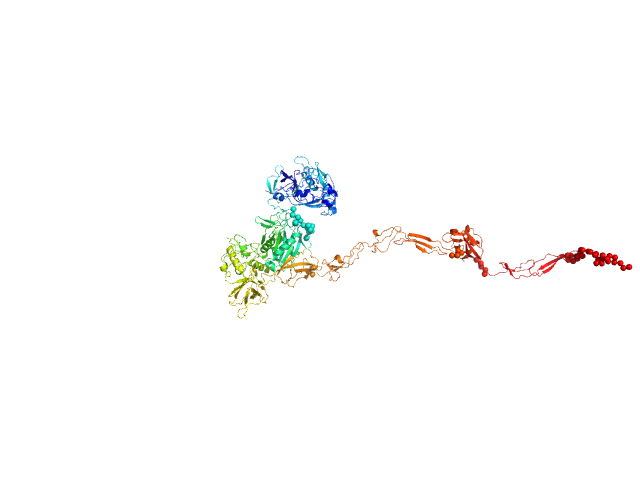
UniProt ID: Q9AES2 (31-1195) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 131 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
5.5 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
UniProt ID: P20711 (1-478) Aromatic-L-amino-acid decarboxylase (L353P)
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (L353P) dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
UniProt ID: P20711 (1-478) Aromatic-L-amino-acid decarboxylase (R347Q)
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (R347Q) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 µM pyridoxal 5'-phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
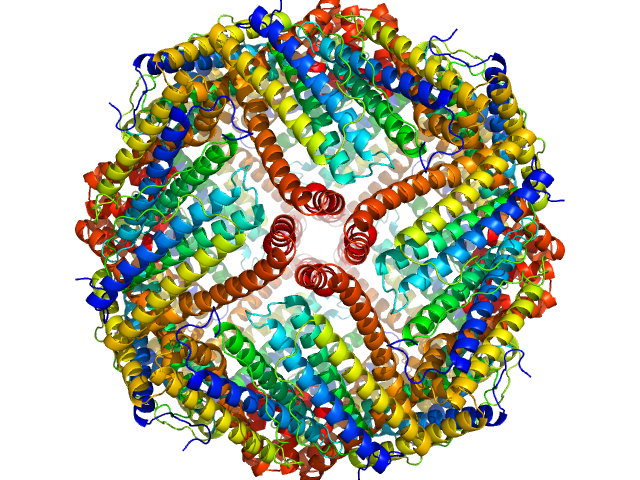
UniProt ID: P02791 (1-175) Ferritin light chain
|
|
|
|
| Sample: |
Ferritin light chain 24-mer, 480 kDa Equus caballus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
5.4 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
632 |
nm3 |
|
|
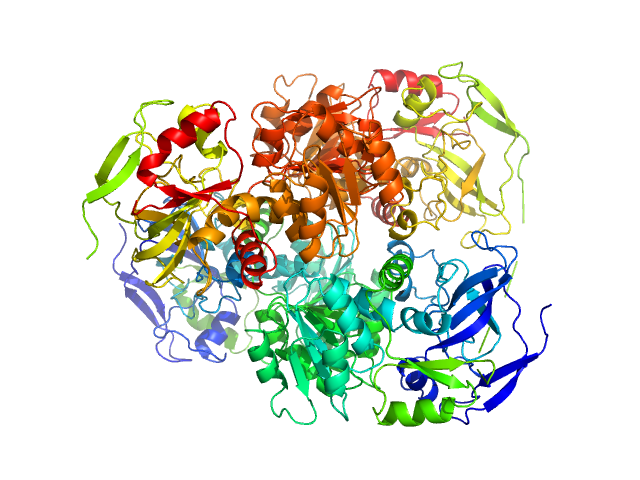
UniProt ID: P00330 (1-348) Alcohol dehydrogenase 1
|
|
|
|
| Sample: |
Alcohol dehydrogenase 1 tetramer, 147 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
197 |
nm3 |
|
|