UniProt ID: P02787 (1-698) Serotransferrin
|
|
|

|
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl (APO Buffer), pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
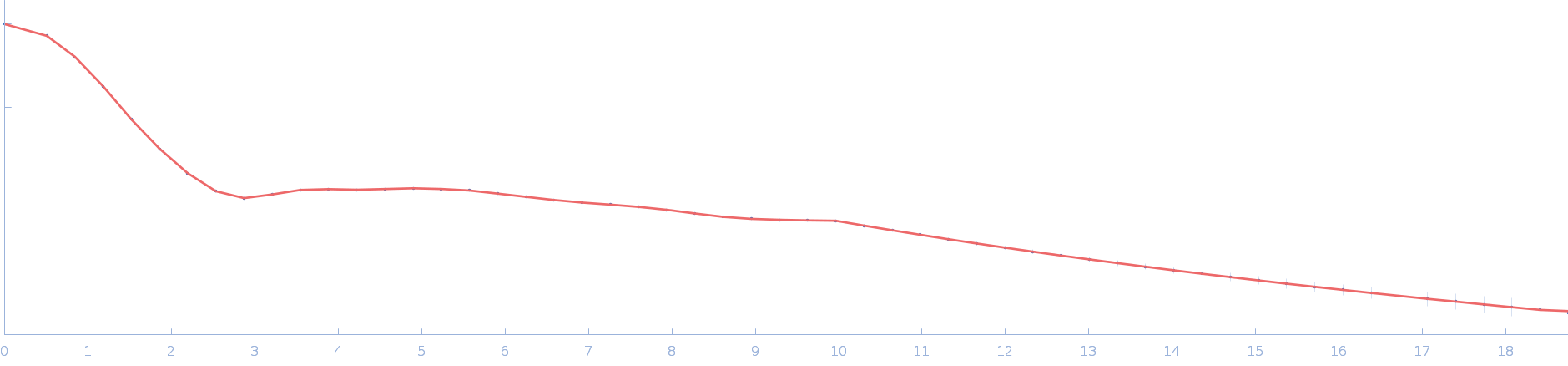
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
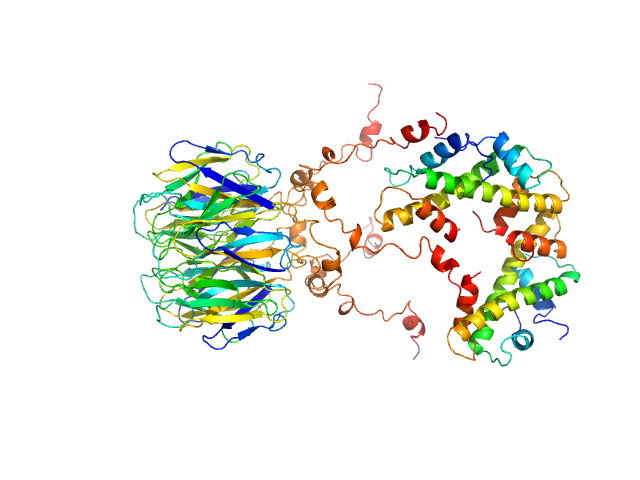
UniProt ID: F4J9Q6 (1-136) Peptidyl-prolyl cis-trans isomerase FKBP43
UniProt ID: P68431 (1-136) Histone H3.1
UniProt ID: P62805 (1-103) Histone H4
|
|
|
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
Histone H3.1 dimer, 31 kDa Homo sapiens protein
Histone H4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
UniProt ID: P74102 (2-317) Orange carotenoid-binding protein
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
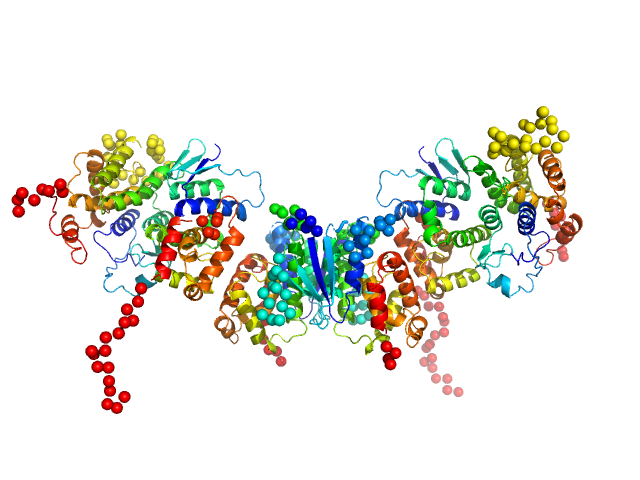
UniProt ID: A0A509AF36 (2-228) Methionine--tRNA ligase
UniProt ID: A0A509ARF0 (2-202) tRNA import protein tRIP
UniProt ID: A0A509AR09 (2-228) Glutamate--tRNA ligase
|
|
|
|
| Sample: |
Methionine--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
UniProt ID: P73129 (1-96) Ssr1698 protein
|
|
|
|
| Sample: |
Ssr1698 protein dimer, 23 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Sep 27
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P54760 (586-602) Ephrin type-B receptor 4
UniProt ID: P20936 (174-1047) Ras GTPase-activating protein 1
|
|
|
|
| Sample: |
Ephrin type-B receptor 4 monomer, 2 kDa Homo sapiens protein
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 4
|
Diverse p120RasGAP interactions with doubly phosphorylated partners EphB4, p190RhoGAP and Dok1
Journal of Biological Chemistry :105098 (2023)
Vish K, Stiegler A, Boggon T
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
176 |
nm3 |
|
|
UniProt ID: Q9NZ71-6 (756-1300) Regulator of telomere elongation helicase 1 (Isoform 6, 756-1300)
|
|
|
|
| Sample: |
Regulator of telomere elongation helicase 1 (Isoform 6, 756-1300) monomer, 59 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl,10 (v/v)% glycerol, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 22
|
Structural and biochemical characterization of the C‐terminal region of the human RTEL
1 helicase
Protein Science 33(9) (2024)
Cortone G, Graewert M, Kanade M, Longo A, Hegde R, González‐Magaña A, Chaves‐Arquero B, Blanco F, Napolitano L, Onesti S
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
UniProt ID: A0A804LC01 (30-200) Protein PAIR1
|
|
|

|
| Sample: |
Protein PAIR1 tetramer, 132 kDa Zea mays protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
8.0 |
nm |
| Dmax |
31.7 |
nm |
|
|
UniProt ID: Q96RR4 (93-517) Calcium/calmodulin-dependent protein kinase kinase 2
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase kinase 2 monomer, 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM TCEP, 3% (w/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
14-3-3 protein inhibits CaMKK1 by blocking the kinase active site with its last two C-terminal helices.
Protein Sci :e4805 (2023)
Petrvalska O, Honzejkova K, Koupilova N, Herman P, Obsilova V, Obsil T
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P0DTC9 (176-245) Nucleoprotein (L223P, L227P, L230P)
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
SARS-CoV-2 N-protein variants: N1-246 and IDL176-246
Guillem Hernandez
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|