UniProt ID: P09651-2 (186-320) Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1
|
|
|
|
| Sample: |
Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 19
|
Design of intrinsically disordered protein variants with diverse structural properties
Science Advances 10(35) (2024)
Pesce F, Bremer A, Tesei G, Hopkins J, Grace C, Mittag T, Lindorff-Larsen K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
UniProt ID: Q9WTS6-1 (343-2715) Isoform A1B1 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
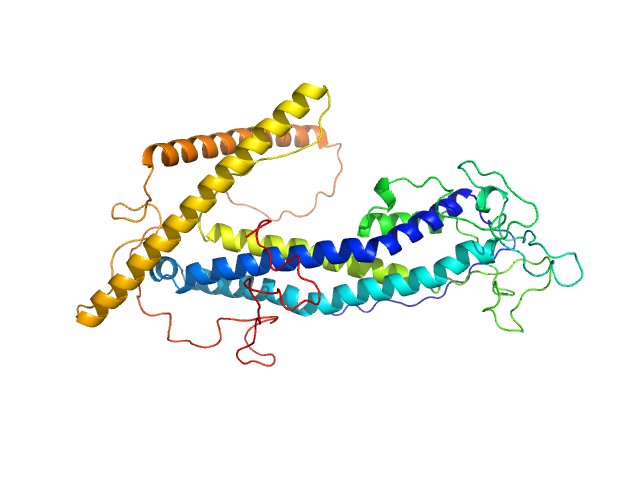
UniProt ID: Q1WK95 (29-462) ISG75
|
|
|
|
| Sample: |
ISG75 monomer, 50 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: J3QLL0 (2-73) Importin subunit alpha-1
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline containing 6 M urea, 0.3 M KCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 29
|
A genetically encoded anomalous SAXS ruler to probe the dimensions of intrinsically disordered proteins.
Proc Natl Acad Sci U S A 121(50):e2415220121 (2024)
Yu M, Gruzinov AY, Ruan H, Scheidt T, Chowdhury A, Giofrè S, Mohammed ASA, Caria J, Sauter PF, Svergun DI, Lemke EA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
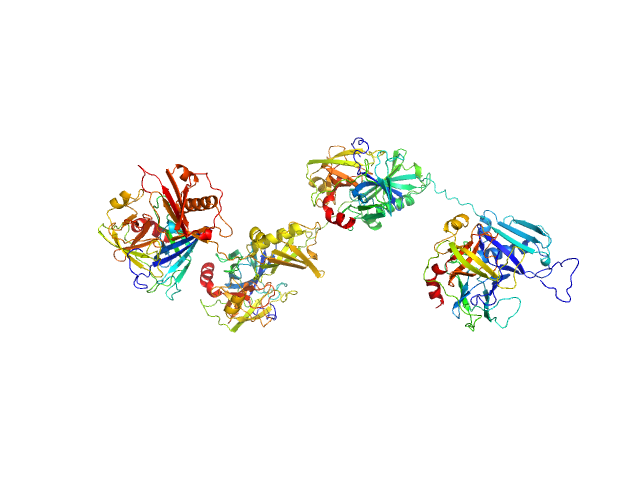
UniProt ID: Q99QS1 (32-478) Protein map
UniProt ID: P08311 (22-245) Cathepsin G
|
|
|
|
| Sample: |
Protein map monomer, 50 kDa Staphylococcus aureus (strain … protein
Cathepsin G tetramer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
UniProt ID: Q8N4Q1 (1-142) Mitochondrial intermembrane space import and assembly protein 40
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: Q9UBC3 (215-853) DNA methyltransferase 3 beta (215-853)
UniProt ID: Q9UJW3 (178-379) DNA methyltransferase 3-like (178-379)
|
|
|

|
| Sample: |
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
271263 |
nm3 |
|
|
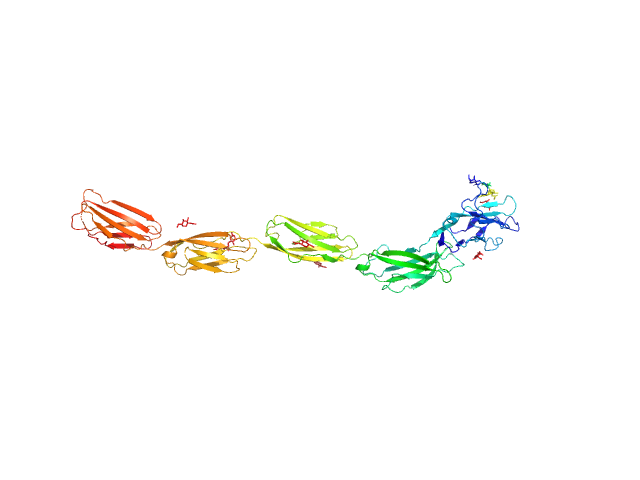
UniProt ID: P20917 (20-508) Myelin-associated glycoprotein (20-508; I473E mutant)
|
|
|
|
| Sample: |
Myelin-associated glycoprotein (20-508; I473E mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: Q46085 (718-1021) Collagenase ColH segement s2as2bs3
|
|
|

|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
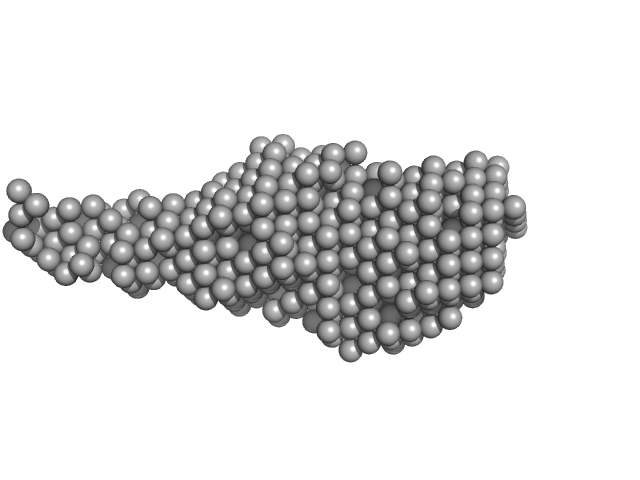
UniProt ID: A0A0T9L251 (None-None) Probable exodeoxyribonuclease III protein XthA
UniProt ID: P9WNV1 (601-691) M. tb. LigA BRCT domain
|
|
|
|
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
M. tb. LigA BRCT domain monomer, 15 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl 5mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 9
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|