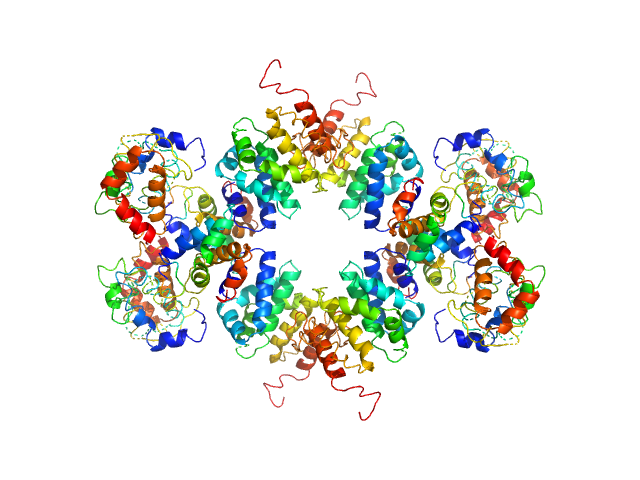
UniProt ID: Q9LM76 (382-801) U-box domain-containing protein 44
|
|
|
|
| Sample: |
U-box domain-containing protein 44 tetramer, 185 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
245 |
nm3 |
|
|
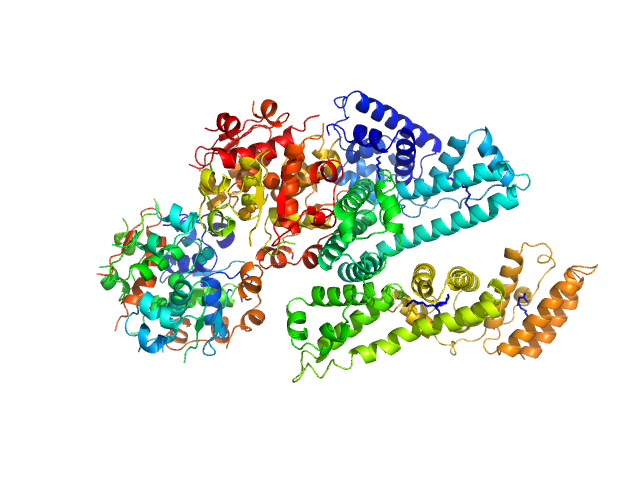
UniProt ID: P02768 (25-609) Human Albumin (Recombumin(R) Elite, Albumedix Ltd.)
UniProt ID: P01308 (25-110) Insulin degludec(Tresiba(R), Novo Nordisk A/S)
|
|
|
|
| Sample: |
Human Albumin (Recombumin(R) Elite, Albumedix Ltd.) monomer, 66 kDa protein
Insulin degludec(Tresiba(R), Novo Nordisk A/S) dodecamer, 73 kDa protein
|
| Buffer: |
25 mM Na2HPO4, 15.9 mM m-cresol, 15.9 mM phenol, 212.8 mM glycerol, 20 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 27
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
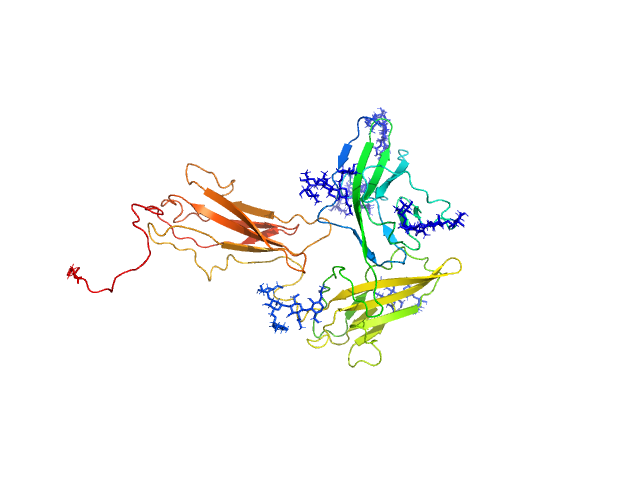
UniProt ID: Q9NPH3 (21-348) Interleukin-1 receptor accessory protein ectodomains with RII linker
|
|
|
|
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with RII linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
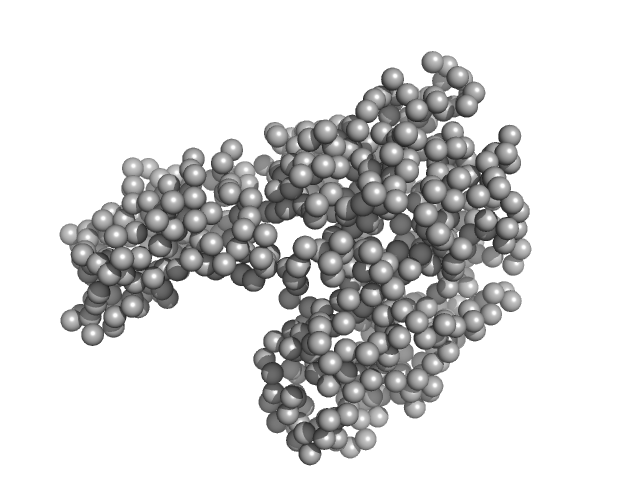
UniProt ID: P02769 (25-607) Bovine serum albumin
|
|
|
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, 3% v/v glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 23
|
Adding Size Exclusion Chromatography (SEC) and Light Scattering (LS) Devices to Obtain High-Quality Small Angle X-Ray Scattering (SAXS) Data
Crystals 10(11):975 (2020)
Graewert M, Da Vela S, Gräwert T, Molodenskiy D, Blanchet C, Svergun D, Jeffries C
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
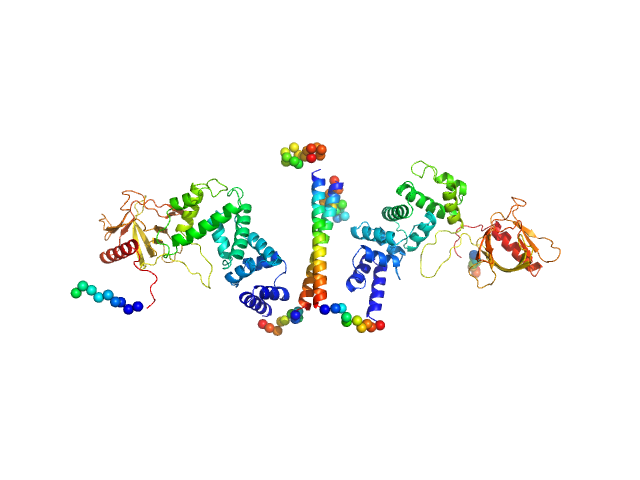
UniProt ID: O08967 (14-390) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
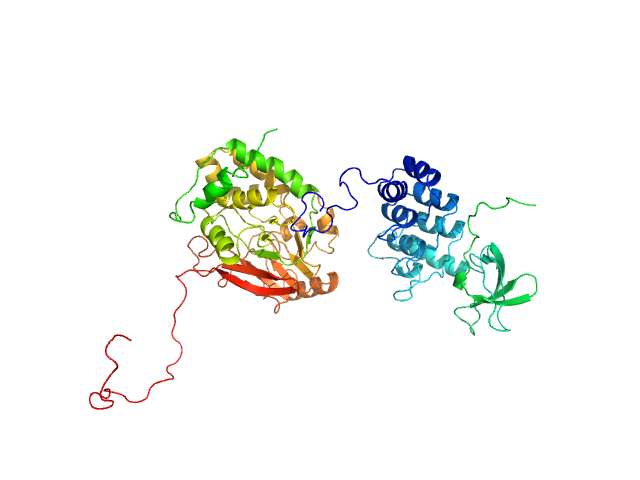
UniProt ID: P62136 (1-330) Serine/threonine-protein phosphatase PP1-alpha catalytic subunit
UniProt ID: Q13625 (905-1128) Apoptosis-stimulating of p53 protein 2
|
|
|
|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Apoptosis-stimulating of p53 protein 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
172 |
nm3 |
|
|
UniProt ID: P52129 (2-357) mRNA endoribonuclease toxin LS - R318A mutant; N-terminal His-tagged
|
|
|
|
| Sample: |
MRNA endoribonuclease toxin LS - R318A mutant; N-terminal His-tagged dimer, 84 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 25
|
Alternative dimerization is required for activity and inhibition of the HEPN ribonuclease RnlA
Nucleic Acids Research 49(12):7164-7178 (2021)
Garcia-Rodriguez G, Charlier D, Wilmaerts D, Michiels J, Loris R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
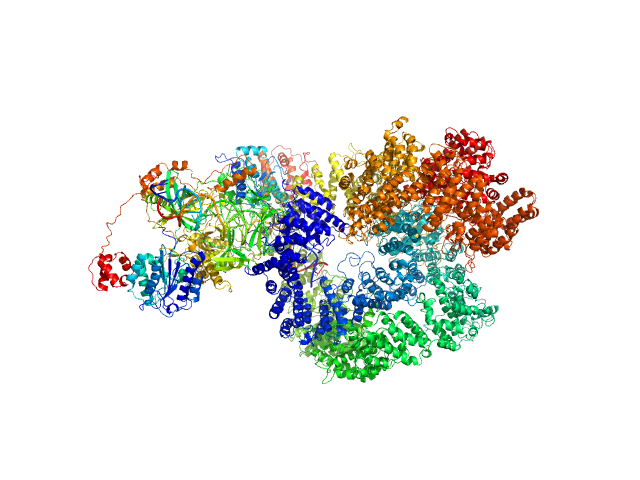
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
UniProt ID: P78527 (10-4128) DNA-dependent protein kinase catalytic subunit
UniProt ID: None (None-None) dsDNA
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
UniProt ID: I6YC99 (36-400) Mce-family protein Mce4A
UniProt ID: None (None-None) n-Dodecyl-β-D-Maltopyranoside
|
|
|

|
| Sample: |
Mce-family protein Mce4A monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 13
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|