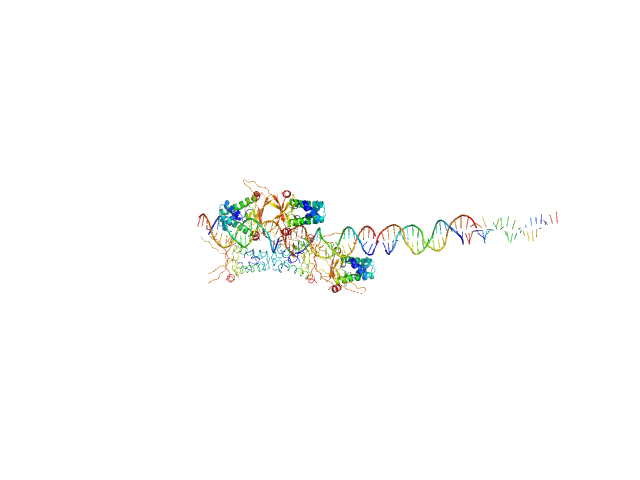
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha decamer, 95 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 300 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
24.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.9 |
nm |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 110 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
5.2 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
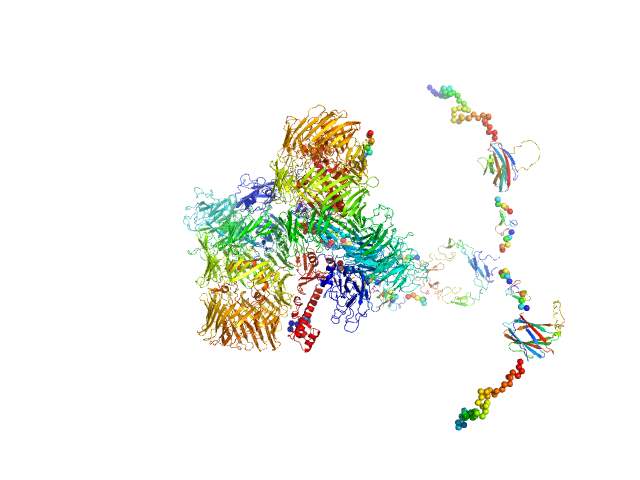
UniProt ID: Q9WTS6-4 (343-2708) Isoform A1B0 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A1B0 of Teneurin-3 dimer, 541 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1089 |
nm3 |
|
|
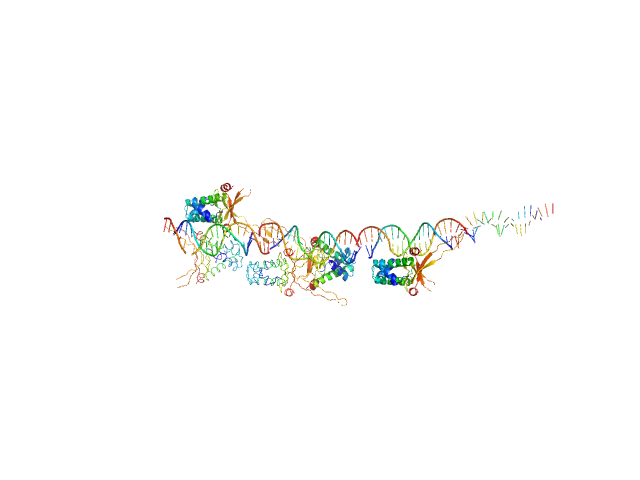
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
8.9 |
nm |
| Dmax |
28.5 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
UniProt ID: Q9WTS6-4 (343-2708) Isoform A1B0 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A1B0 of Teneurin-3 dimer, 541 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.7 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1092 |
nm3 |
|
|
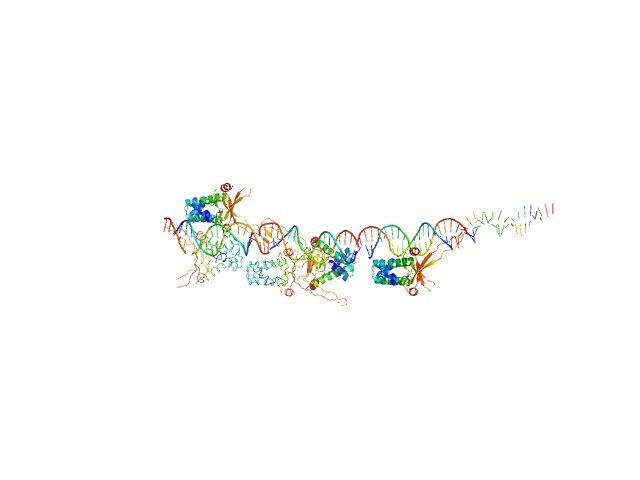
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Nucleic Acids Res 53(11) (2025)
Hernandez G, Martins ML, Fernandes NP, Veloso T, Lopes J, Gomes T, Cordeiro TN
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
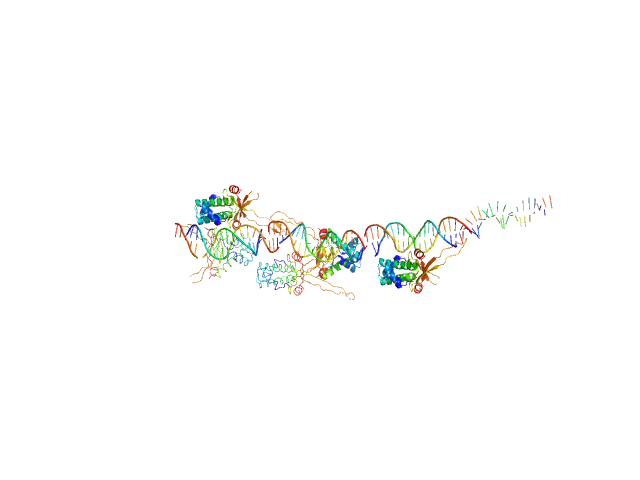
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 14-mer, 133 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
5.8 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
308 |
nm3 |
|
|