UniProt ID: Q73FL6 (65-252) DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein)
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) monomer, 21 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
UniProt ID: O08967 (63-399) Cytohesin-3
|
|
|

|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 13
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
UniProt ID: O08967 (63-399) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.001 mM inositol 1,3,4,5-tetrakisphosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Mar 21
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
UniProt ID: Q73FL6 (16-252) DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein)
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) trimer, 81 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2014 Mar 29
|
The atypical thiol-disulfide exchange protein α-DsbA2 from Wolbachia pipientis is a homotrimeric disulfide isomerase.
Acta Crystallogr D Struct Biol 75(Pt 3):283-295 (2019)
Walden PM, Whitten AE, Premkumar L, Halili MA, Heras B, King GJ, Martin JL
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
913 |
nm3 |
|
|
UniProt ID: Q73FL6 (65-252) DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein)
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) monomer, 21 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Feb 29
|
The atypical thiol-disulfide exchange protein α-DsbA2 from Wolbachia pipientis is a homotrimeric disulfide isomerase.
Acta Crystallogr D Struct Biol 75(Pt 3):283-295 (2019)
Walden PM, Whitten AE, Premkumar L, Halili MA, Heras B, King GJ, Martin JL
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
UniProt ID: C2LPE2 (22-243) DsbA-like protein
UniProt ID: B4EV20 (22-275) Putative metal resistance protein
|
|
|
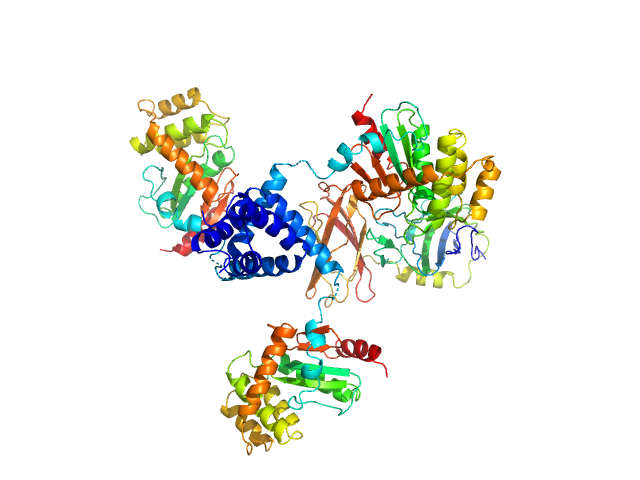
|
| Sample: |
DsbA-like protein trimer, 74 kDa Proteus mirabilis protein
Putative metal resistance protein monomer, 30 kDa Proteus mirabilis protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 2
|
Disulfide isomerase activity of the dynamic, trimeric Proteus mirabilis ScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J Biol Chem 293(16):5793-5805 (2018)
Furlong EJ, Choudhury HG, Kurth F, Duff AP, Whitten AE, Martin JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
UniProt ID: Q7MX92 (None-None) Peptidase, M49 family
|
|
|
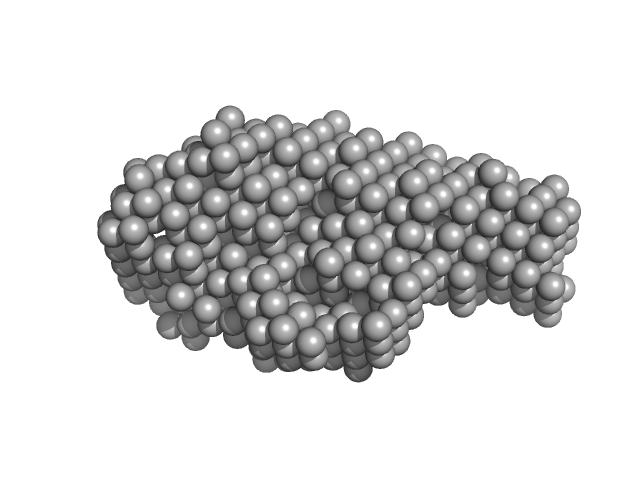
|
| Sample: |
Peptidase, M49 family monomer, 102 kDa Porphyromonas gingivalis protein
|
| Buffer: |
50mM Tris,100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 14
|
A novel Porphyromonas gingivalis enzyme: An atypical dipeptidyl peptidase III with an ARM repeat domain.
PLoS One 12(11):e0188915 (2017)
Hromić-Jahjefendić A, Jajčanin Jozić N, Kazazić S, Grabar Branilović M, Karačić Z, Schrittwieser JH, Das KMP, Tomin M, Oberer M, Gruber K, Abramić M, Tomić S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
UniProt ID: P40699 (203-303) Surface presentation of antigens protein SpaO SpaO(SPOA2)
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
UniProt ID: P40699 (203-303) Surface presentation of antigens protein SpaO SpaO(SPOA2)
UniProt ID: P40699 (1-303) Surface presentation of antigens protein SpaO(SPOA1,2)
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) monomer, 34 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
UniProt ID: P40699 (1-145) Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus monomer, 17 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|