UniProt ID: P33316 (94-252) Deoxyuridine 5'-triphosphate nucleotidohydrolase
UniProt ID: E2FZP6 (13-278) SaPIbov1 pathogenicity island repressor
|
|
|

|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
UniProt ID: P49333 (339-738) Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding + receiver domains
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding + receiver domains dimer, 88 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
UniProt ID: P49333 (339-589) Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding domains
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding domains dimer, 55 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: A0A0H2ZNP2 (None-None) DHH subfamily 1 protein
|
|
|
|
| Sample: |
DHH subfamily 1 protein dimer, 70 kDa Streptococcus pneumoniae serotype … protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: Q9X1T1 (None-None) T.maritima PDE
|
|
|
|
| Sample: |
T.maritima PDE dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
25mM Tris 500mM NaCl 3% (v/v) glycerol 2mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 17
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
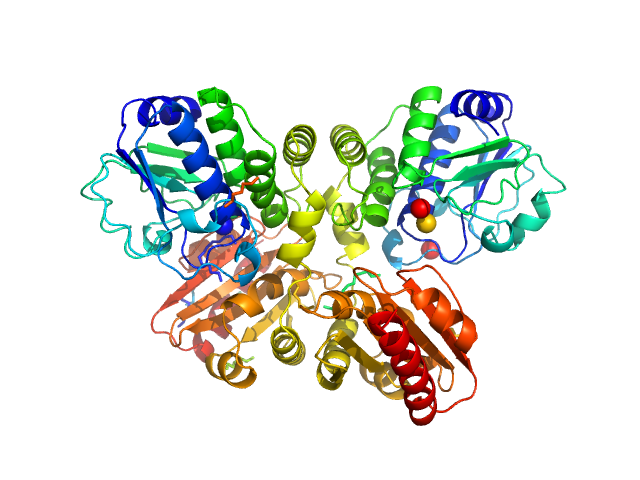
UniProt ID: Q9X1T1 (None-None) Thermotoga maritima phosphodiesterase (wildtype, TmPDE, TM1595)
|
|
|
|
| Sample: |
Thermotoga maritima phosphodiesterase (wildtype, TmPDE, TM1595) dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
UniProt ID: Q6PHU5 (None-None) Sortilin, also: Neurotensin-receptor 3
|
|
|
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
UniProt ID: Q6PHU5 (None-None) Sortilin, also: Neurotensin-receptor 3
|
|
|
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
UniProt ID: P0CG48 (305-379) Polyubiquitin-C
|
|
|
|
| Sample: |
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Mar 24
|
Characterizing Protein Dynamics with Integrative Use of Bulk and Single-Molecule Techniques.
Biochemistry 57(3):305-313 (2018)
Liu Z, Gong Z, Cao Y, Ding YH, Dong MQ, Lu YB, Zhang WP, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: Q9JTL9 (23-146) N-terminus of disulfide interchange protein DsbD
|
|
|

|
| Sample: |
N-terminus of disulfide interchange protein DsbD monomer, 14 kDa Neisseria meningitidis protein
|
| Buffer: |
25mM HEPES 150mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Production, biophysical characterization and initial crystallization studies of the N- and C-terminal domains of DsbD, an essential enzyme in Neisseria meningitidis.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):31-38 (2018)
Smith RP, Whitten AE, Paxman JJ, Kahler CM, Scanlon MJ, Heras B
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
17 |
nm3 |
|
|