UniProt ID: P40699 (203-303) Surface presentation of antigens protein SpaO SpaO(SPOA2)
UniProt ID: P40699 (1-145) Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) N-terminus monomer, 17 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
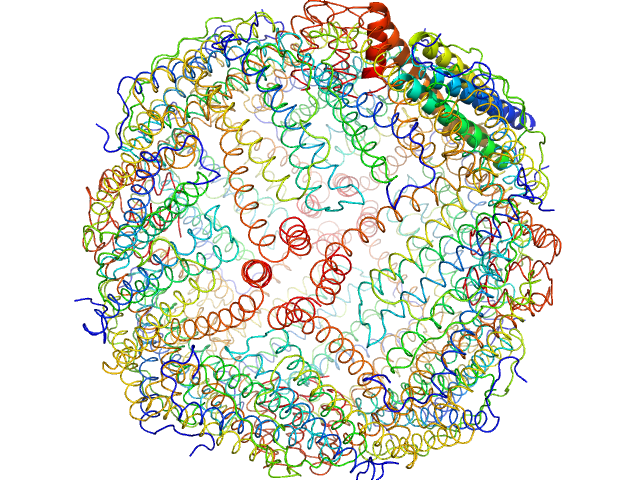
UniProt ID: P02791 (None-None) Horse spleen apoferritin
|
|
|
|
| Sample: |
Horse spleen apoferritin 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
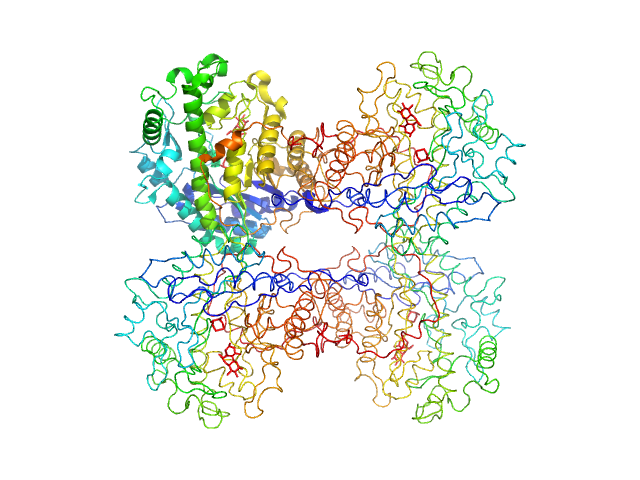
UniProt ID: P10537 (None-None) Beta-amylase
|
|
|
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
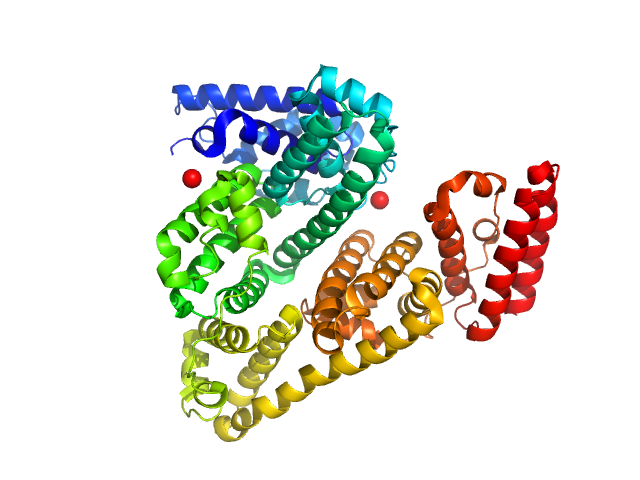
UniProt ID: P02769 (25-607) Serum albumin
|
|
|
|
| Sample: |
Serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
Hepes, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 25
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
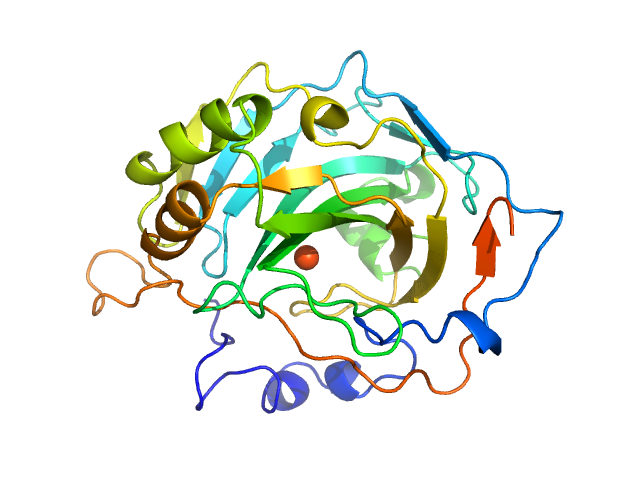
UniProt ID: P00921 (None-None) Carbonic anhydrase 2
|
|
|
|
| Sample: |
Carbonic anhydrase 2 monomer, 29 kDa Bos taurus protein
|
| Buffer: |
Tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 16
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
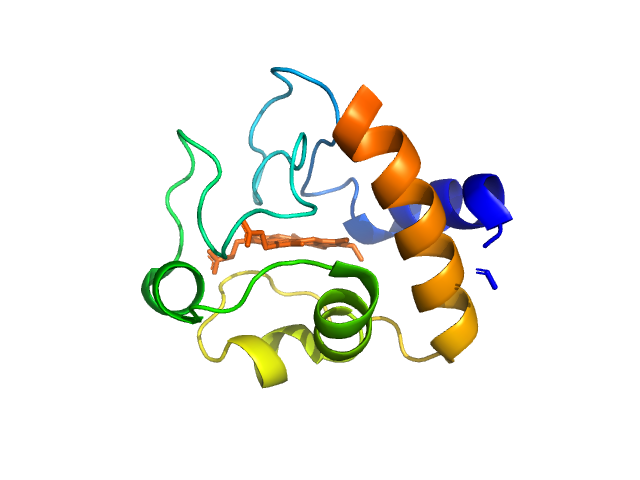
UniProt ID: P00004 (None-None) Cytochrome c
|
|
|
|
| Sample: |
Cytochrome c monomer, 12 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 16
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
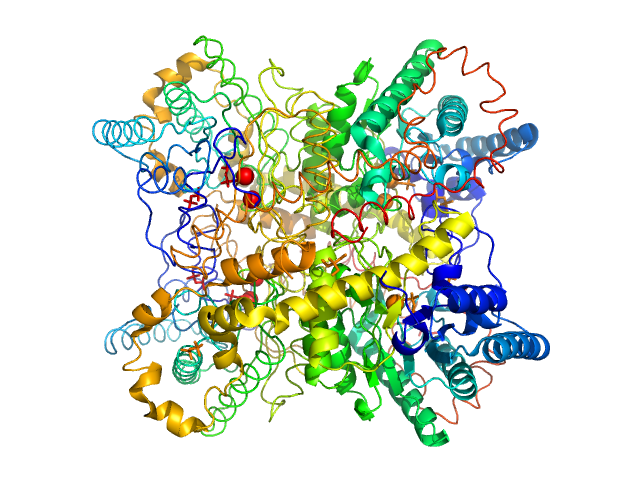
UniProt ID: P24300 (None-None) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 172 kDa Streptomyces rubiginosus protein
|
| Buffer: |
100 mM tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 17
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
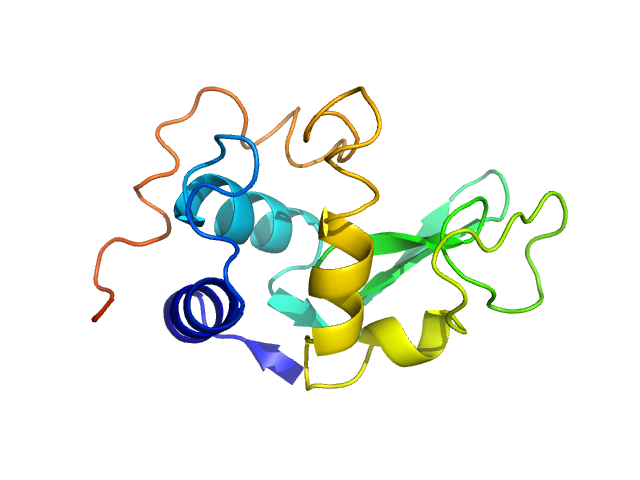
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM Sodium Acetate pH 4.0, 50 mM NaCl, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 May 31
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
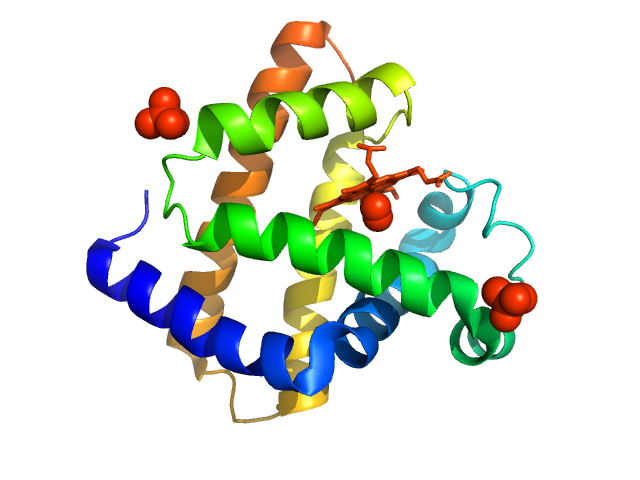
UniProt ID: P68082 (None-None) Myoglobin
|
|
|
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
100 mM tris pH 7.5, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
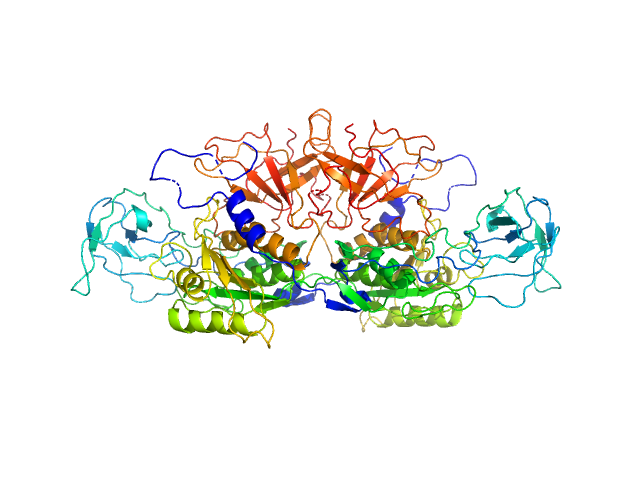
UniProt ID: Q39054 (1-451) Molybdopterin biosynthesis protein CNX1
|
|
|
|
| Sample: |
Molybdopterin biosynthesis protein CNX1 dimer, 99 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Hepes/KOH, 150 mM NaCl, 1 mM EDTA, 2 % (v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Feb 5
|
Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor.
Biochem J 474(1):163-178 (2017)
Krausze J, Probst C, Curth U, Reichelt J, Saha S, Schafflick D, Heinz DW, Mendel RR, Kruse T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|