UniProt ID: O53943 (None-None) EspG5 chaperone from Mycobacterium tuberculosis
UniProt ID: None (None-None) PE25 from Mycobacterium tuberculosis
UniProt ID: Q79FE1 (None-None) PPE41 from Mycobacterium tuberculosis
|
|
|
|
| Sample: |
EspG5 chaperone from Mycobacterium tuberculosis monomer, 32 kDa Mycobacterium tuberculosis protein
PE25 from Mycobacterium tuberculosis monomer, 11 kDa Mycobacterium tuberculosis protein
PPE41 from Mycobacterium tuberculosis monomer, 22 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Feb 10
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
|
|
UniProt ID: P9WJC6 (None-None) EspG3 chaperone from Mycobacterium tuberculosis
UniProt ID: L7N695 (None-None) PE5 from Mycobacterium tuberculosis
UniProt ID: P9WI43 (None-None) PPE4 from Mycobacterium tuberculosis
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium tuberculosis, 34 kDa Mycobacterium tuberculosis protein
PE5 from Mycobacterium tuberculosis monomer, 10 kDa Mycobacterium tuberculosis protein
PPE4 from Mycobacterium tuberculosis monomer, 52 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jun 15
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.2 |
nm |
|
|
UniProt ID: Q80XU8 (17-371) Leucine-rich repeat and fibronectin type-III domain-containing protein 4
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 79 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
UniProt ID: Q80XU8 (17-510) Leucine-rich repeat and fibronectin type-III domain-containing protein 4
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 109 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 11
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
313 |
nm3 |
|
|
UniProt ID: Q8BXA0 (18-376) Leucine-rich repeat and fibronectin type-III domain-containing protein 5
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 5 dimer, 82 kDa Mus musculus protein
|
| Buffer: |
30 mM Tris-Cl, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jun 8
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
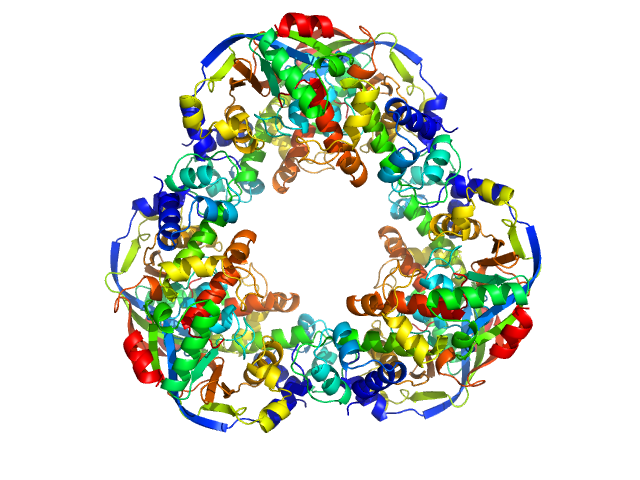
UniProt ID: P9WLP9 (None-None) Mycobacterial cidal toxin
UniProt ID: P9WLP7 (None-None) Mycobacterial cidal antitoxin
|
|
|
|
| Sample: |
Mycobacterial cidal toxin hexamer, 121 kDa Mycobacterium tuberculosis protein
Mycobacterial cidal antitoxin hexamer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
100 mM HEPES, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
An NAD+ Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol Cell (2019)
Freire DM, Gutierrez C, Garza-Garcia A, Grabowska AD, Sala AJ, Ariyachaokun K, Panikova T, Beckham KSH, Colom A, Pogenberg V, Cianci M, Tuukkanen A, Boudehen YM, Peixoto A, Botella L, Svergun DI, Schnappinger D, Schneider TR, Genevaux P, de Carvalho LPS, Wilmanns M, Parret AHA, Neyrolles O
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
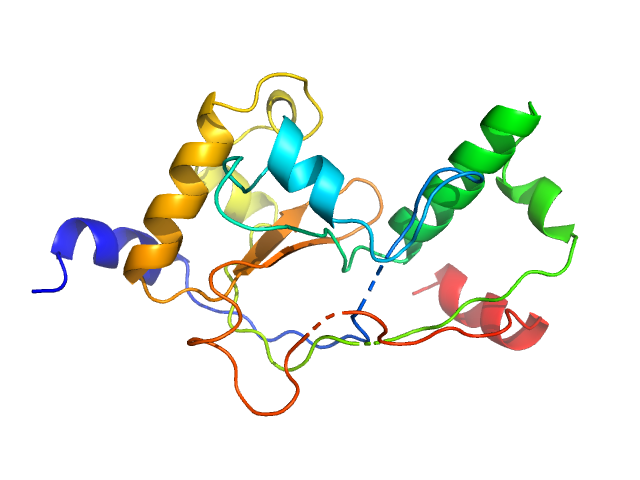
UniProt ID: P9WLP9 (1-186) Mycobacterial cidal toxin
|
|
|
|
| Sample: |
Mycobacterial cidal toxin monomer, 20 kDa Mycobacterium tuberculosis H37Rv protein
|
| Buffer: |
30 mM Tris-HCl, 200 mM NaCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 2
|
MbsTA
Diana Freire
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
UniProt ID: K0J1W8 (24-664) Bacterial cellulose synthesis subunit C
|
|
|
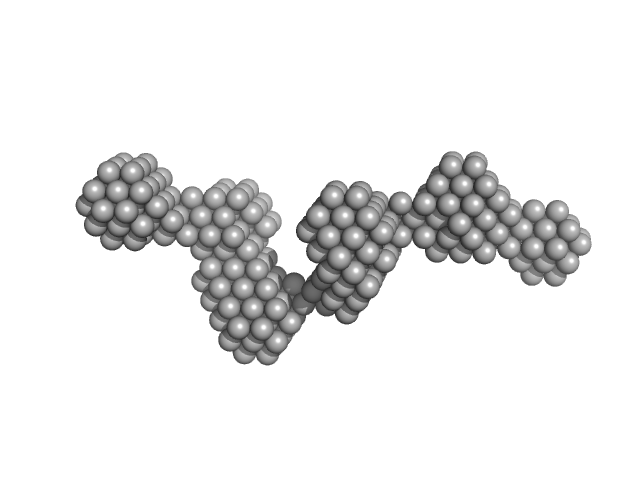
|
| Sample: |
Bacterial cellulose synthesis subunit C monomer, 71 kDa Enterobacter sp. CJF-002 protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Apr 24
|
Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C.
Sci Rep 7(1):13018 (2017)
Nojima S, Fujishima A, Kato K, Ohuchi K, Shimizu N, Yonezawa K, Tajima K, Yao M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
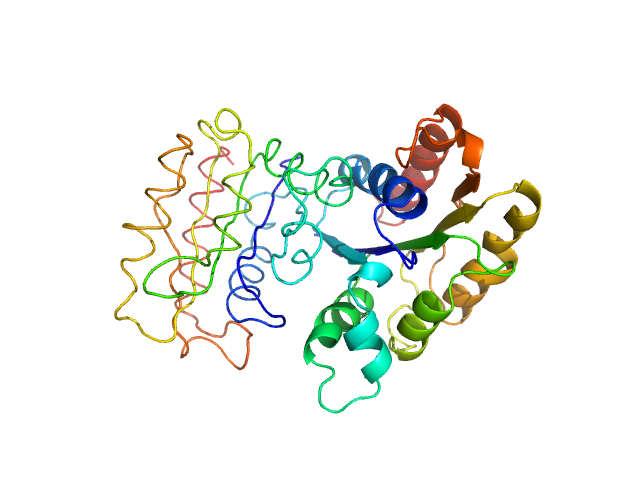
UniProt ID: A1BCD5 (None-None) NADPH-dependent FMN reductase
|
|
|
|
| Sample: |
NADPH-dependent FMN reductase dimer, 41 kDa Paracoccus denitrificans protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Feb 22
|
Structural Insight into Catalysis by the Flavin-Dependent NADH Oxidase (Pden_5119) of Paracoccus denitrificans.
Int J Mol Sci 24(4) (2023)
Kryl M, Sedláček V, Kučera I
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P05090 (21-189) Apolipoprotein D
|
|
|
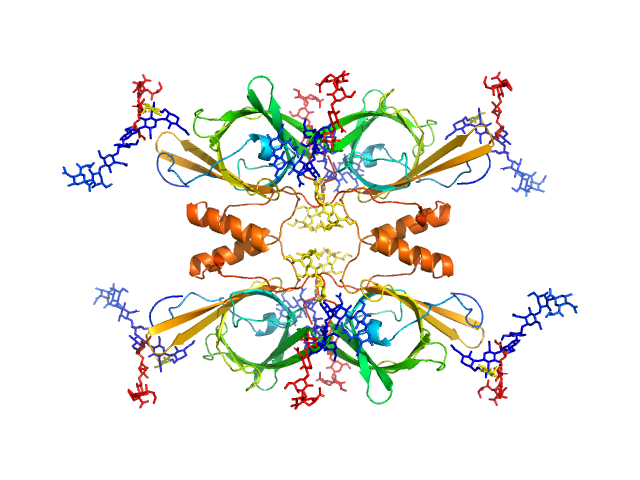
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Identification of a novel tetrameric structure for human apolipoprotein-D.
J Struct Biol 203(3):205-218 (2018)
Kielkopf CS, Low JKK, Mok YF, Bhatia S, Palasovski T, Oakley AJ, Whitten AE, Garner B, Brown SHJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
169 |
nm3 |
|
|