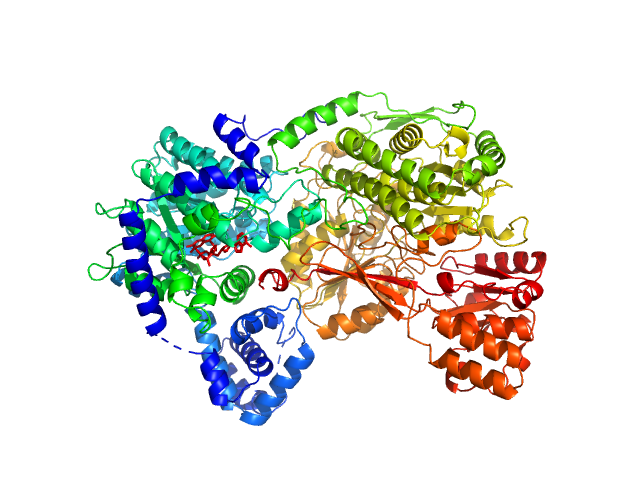
UniProt ID: F7X6I3 (None-None) Sinorhizobium meliloti (SmPutA)
|
|
|
|
| Sample: |
Sinorhizobium meliloti (SmPutA) monomer, 132 kDa Sinorhizobium meliloti protein
|
| Buffer: |
50 mM Tris, 1% (v/v) glycerol, 0.5 mM THP, and 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 27
|
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.
J Biol Chem 291(46):24065-24075 (2016)
Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
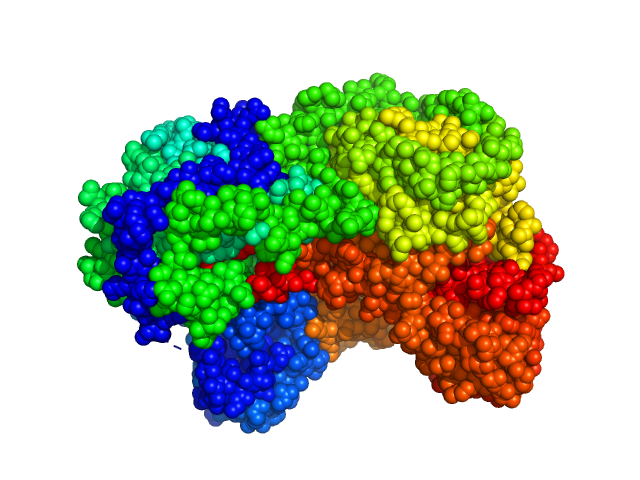
UniProt ID: F7X6I3 (None-None) Sinorhizobium meliloti (SmPutA)
|
|
|
|
| Sample: |
Sinorhizobium meliloti (SmPutA) monomer, 132 kDa Sinorhizobium meliloti protein
|
| Buffer: |
50 mM Tris, 1% (v/v) glycerol, 0.5 mM THP, and 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 27
|
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.
J Biol Chem 291(46):24065-24075 (2016)
Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
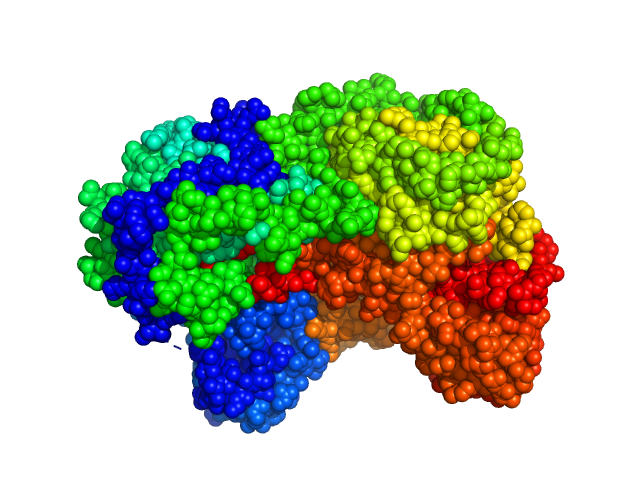
UniProt ID: F7X6I3 (None-None) Sinorhizobium meliloti (SmPutA)
|
|
|
|
| Sample: |
Sinorhizobium meliloti (SmPutA) monomer, 132 kDa Sinorhizobium meliloti protein
|
| Buffer: |
50 mM Tris, 1% (v/v) glycerol, 0.5 mM THP, and 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 27
|
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.
J Biol Chem 291(46):24065-24075 (2016)
Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
248 |
nm3 |
|
|
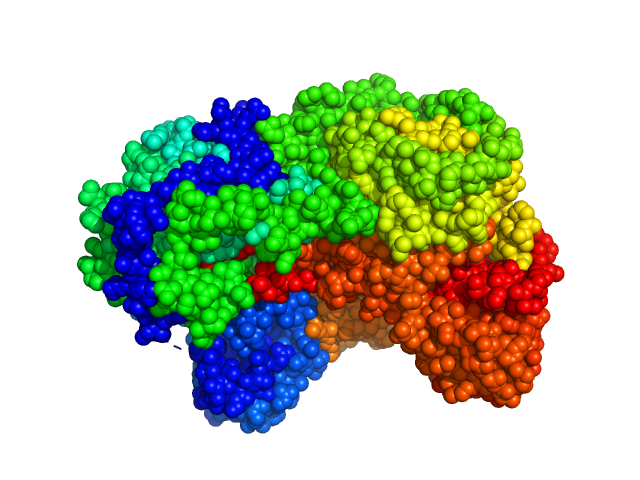
UniProt ID: F7X6I3 (None-None) Sinorhizobium meliloti (SmPutA)
|
|
|
|
| Sample: |
Sinorhizobium meliloti (SmPutA) monomer, 132 kDa Sinorhizobium meliloti protein
|
| Buffer: |
50 mM Tris, 1% (v/v) glycerol, 0.5 mM THP, and 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 27
|
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.
J Biol Chem 291(46):24065-24075 (2016)
Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
UniProt ID: B2HNX0 (None-None) EspG3 chaperone from Mycobacterium marinum M
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium marinum M monomer, 32 kDa Mycobacterium marinum M protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
|
|
UniProt ID: B2HMS9 (None-None) EspG1 from Mycobacterium marinum
|
|
|
|
| Sample: |
EspG1 from Mycobacterium marinum monomer, 30 kDa Mycobacterium marinum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
|
|
UniProt ID: A0QQ45 (None-None) EspG3 chaperone from Mycobacterium smegmatis
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
|
|
UniProt ID: P9WJC6 (None-None) EspG3 chaperone from Mycobacterium tuberculosis
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium tuberculosis, 34 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
|
|
UniProt ID: A0QQ45 (None-None) EspG3 chaperone from Mycobacterium smegmatis
|
|
|
|
| Sample: |
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
|
|
UniProt ID: O53943 (None-None) EspG5 chaperone from Mycobacterium tuberculosis
|
|
|
|
| Sample: |
EspG5 chaperone from Mycobacterium tuberculosis monomer, 32 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 12
|
Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
(2018)
Tuukkanen A, Freire D, Chan S, Arbing M, Reed R, Evans T, Zenkeviciutė G, Kim J, Kahng S, Sawaya M, Chaton C, Wilmanns M, Eisenberg D, Parret A, Korotkov K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
|
|