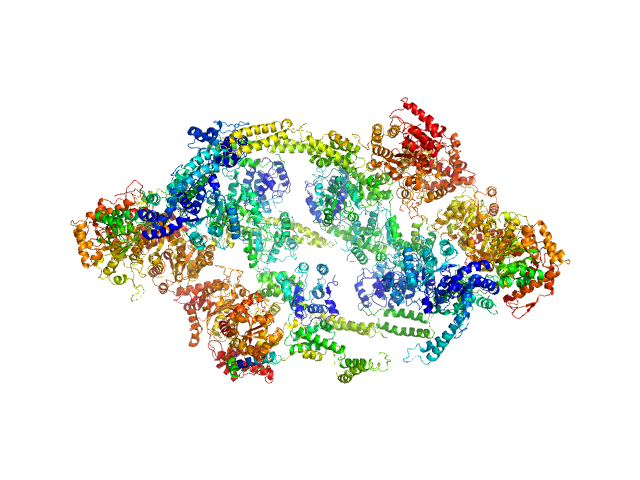
UniProt ID: P9WPC9 (None-None) ATP-dependent Clp protease ATP-binding subunit ClpC1
|
|
|
|
| Sample: |
ATP-dependent Clp protease ATP-binding subunit ClpC1, 95 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
Hepes 50 mM pH 7.5, KCl 100 mM, glycerol 10%, MgCl2 4 mM and ATP 1 mM, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 18
|
The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis.
J Biol Chem 293(22):8379-8393 (2018)
Weinhäupl K, Brennich M, Kazmaier U, Lelievre J, Ballell L, Goldberg A, Schanda P, Fraga H
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2156 |
nm3 |
|
|
UniProt ID: P9WPC9 (None-None) ATP-dependent Clp protease ATP-binding subunit ClpC1
|
|
|
|
| Sample: |
ATP-dependent Clp protease ATP-binding subunit ClpC1, 95 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
Hepes 50 mM pH 7.5, KCl 100 mM, glycerol 10%, MgCl2 4 mM and ATP 1 mM, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 18
|
The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis.
J Biol Chem 293(22):8379-8393 (2018)
Weinhäupl K, Brennich M, Kazmaier U, Lelievre J, Ballell L, Goldberg A, Schanda P, Fraga H
|
| RgGuinier |
7.9 |
nm |
| Dmax |
25.1 |
nm |
| VolumePorod |
2416 |
nm3 |
|
|
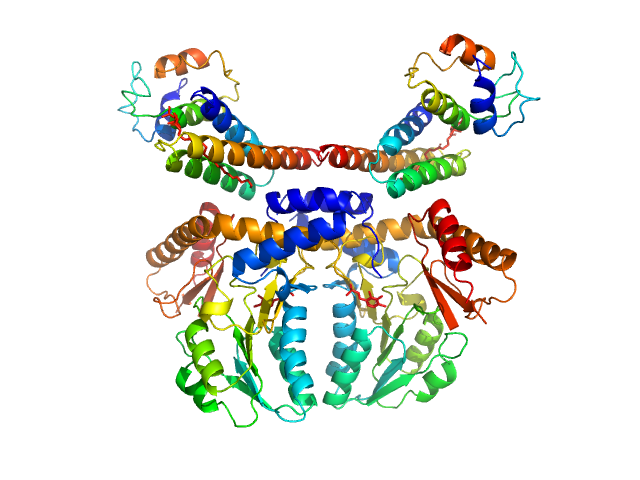
UniProt ID: Q9Y697 (53-457) Cysteine desulfurase, mitochondrial
UniProt ID: Q9HD34 (None-None) LYR motif-containing protein 4
UniProt ID: P0A6A8 (None-None) Acyl carrier protein
|
|
|
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
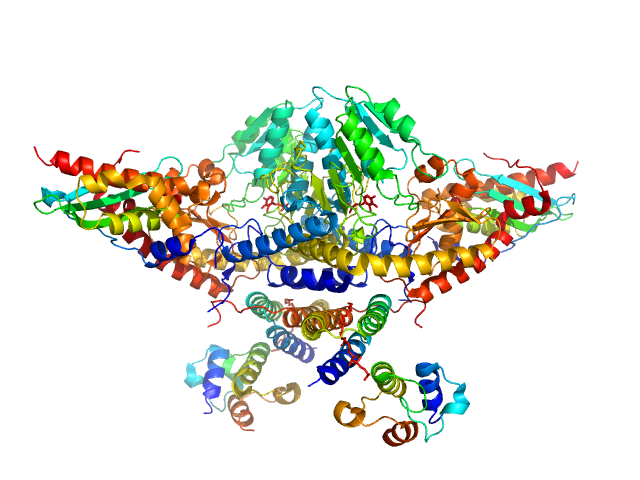
UniProt ID: Q9Y697 (53-457) Cysteine desulfurase, mitochondrial
UniProt ID: Q9HD34 (None-None) LYR motif-containing protein 4
UniProt ID: P0A6A8 (None-None) Acyl carrier protein
UniProt ID: Q9H1K1 (33-167) Iron-sulfur cluster assembly enzyme ISCU, mitochondrial
|
|
|
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
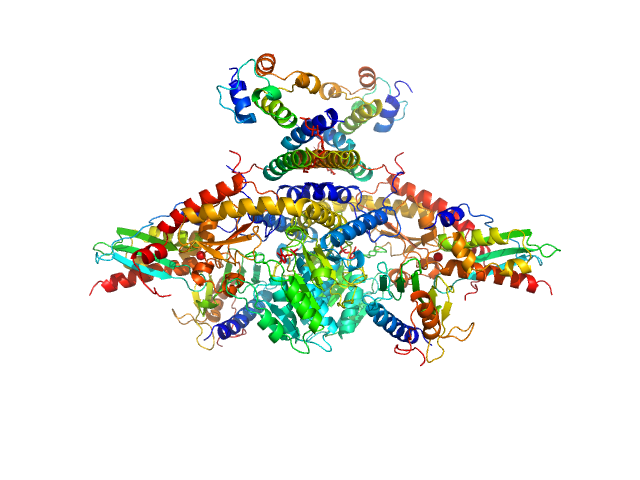
UniProt ID: Q9Y697 (53-457) Cysteine desulfurase, mitochondrial
UniProt ID: Q9HD34 (None-None) LYR motif-containing protein 4
UniProt ID: P0A6A8 (None-None) Acyl carrier protein
UniProt ID: Q9H1K1 (33-167) Iron-sulfur cluster assembly enzyme ISCU, mitochondrial
UniProt ID: Q16595 (81-210) Frataxin, mitochondrial
|
|
|
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
Frataxin, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 Apr 18
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
287 |
nm3 |
|
|
UniProt ID: Q4KRW3 (73-194) Human respiratory syncytial virus M2-1
|
|
|

|
| Sample: |
Human respiratory syncytial virus M2-1 monomer, 14 kDa Human orthopneumovirus protein
|
| Buffer: |
20 mM Tris–HCl, 300 mM NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 13
|
Structure and stability of the Human respiratory syncytial virus M2-1 RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun 74(Pt 1):23-30 (2018)
Molina IG, Josts I, Almeida Hernandez Y, Esperante S, Salgueiro M, Garcia Alai MM, de Prat-Gay G, Tidow H
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
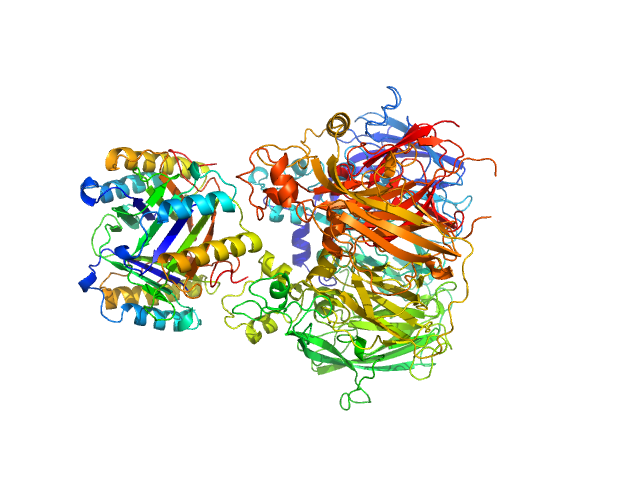
UniProt ID: P14174 (1-115) Macrophage migration inhibitory factor
UniProt ID: P00450 (1-1065) Ceruloplasmin
|
|
|
|
| Sample: |
Macrophage migration inhibitory factor trimer, 37 kDa Homo sapiens protein
Ceruloplasmin monomer, 122 kDa Homo sapiens protein
|
| Buffer: |
50mkm CuSO4, 100mM Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at HECUS System-3, None on 2015 Dec 22
|
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
Biochemistry (Mosc) 83(6):701-707 (2018)
Sokolov AV, Dadinova LA, Petoukhov MV, Bourenkov G, Dubova KM, Amarantov SV, Volkov VV, Kostevich VA, Gorbunov NP, Grudinina NA, Vasilyev VB, Samygina VR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
UniProt ID: None (None-None) Sa0446 binding sequence 40bp
UniProt ID: Q4J9G1 (1-123) Transcriptional regulator Lrs14-like protein
|
|
|
|
| Sample: |
Sa0446 binding sequence 40bp monomer, 25 kDa DNA
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 5
|
Solution Structure of Archaeal Biofilm Regulator 2 (AbfR2) in Complex with 40 bp DNA
Marian Vogt
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
UniProt ID: Q4J9G1 (1-123) Transcriptional regulator Lrs14-like protein
|
|
|

|
| Sample: |
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 May 5
|
Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol 74(Pt 11):1105-1114 (2018)
Vogt MS, Völpel SL, Albers SV, Essen LO, Banerjee A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: P41365 (26-342) Lipase B from Pseudozyma antarctica
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica, 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|