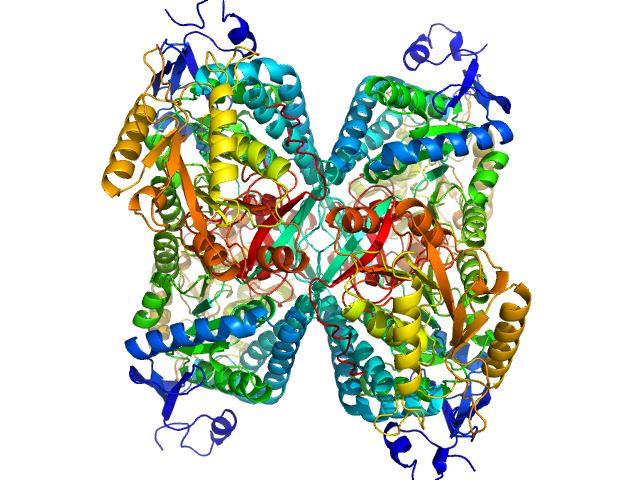
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
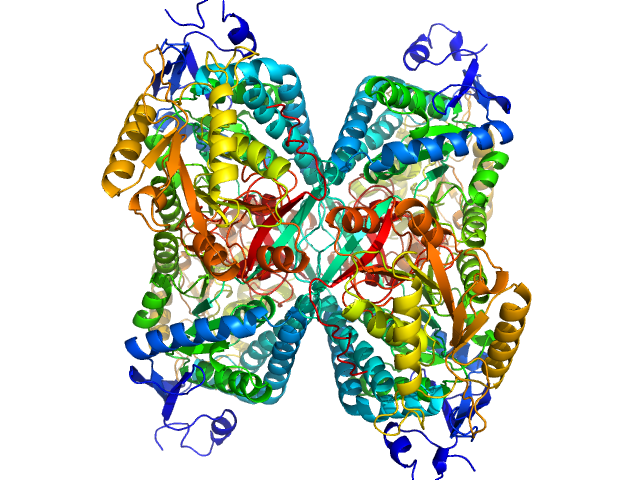
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
UniProt ID: Q9HSF4 (1-116) VNG0258H/RosR
|
|
|

|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
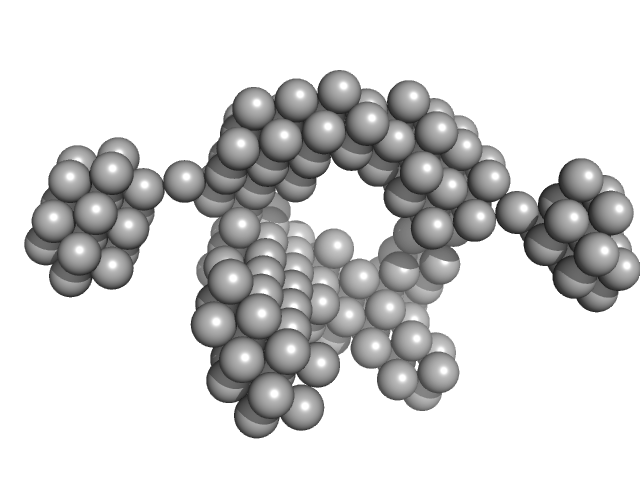
UniProt ID: Q9HSF4 (1-116) VNG0258H/RosR
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
UniProt ID: Q9HSF4 (None-None) VNG0258H/RosR
|
|
|
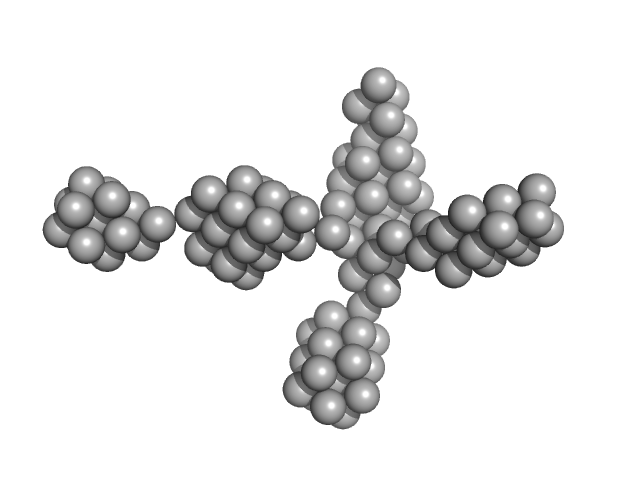
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
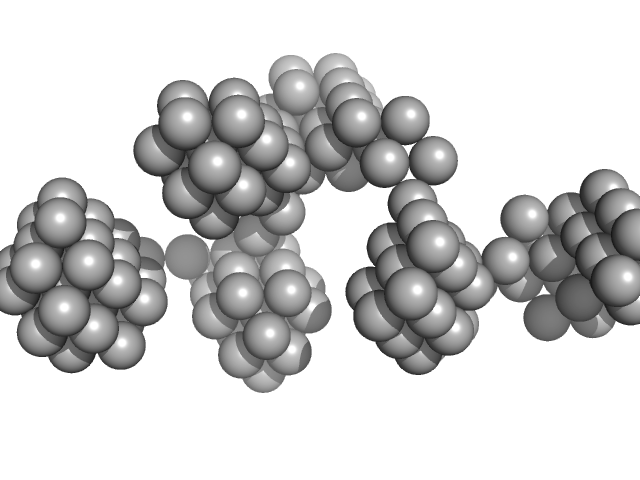
UniProt ID: Q9HSF4 (1-116) VNG0258H/RosR
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 26
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
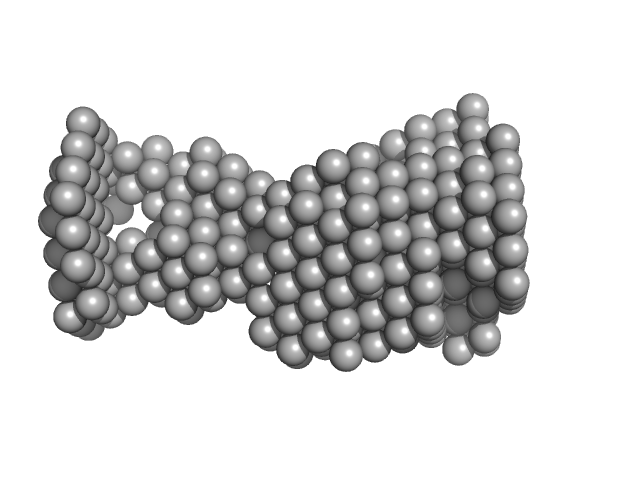
UniProt ID: Q9HSF4 (1-116) VNG0258H/RosR
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M RbCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: Q14896 (319-451) cardiac myosin binding protein C: tri-helix bundle-C2
|
|
|
|
| Sample: |
Cardiac myosin binding protein C: tri-helix bundle-C2 monomer, 15 kDa human sequence obtained … protein
|
| Buffer: |
150 mM NaCl, 10 mM MES, 2 mM TCEP, 1 mM NaN3 at 4°C, pH: 6.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 18
|
A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure 24(11):2000-2007 (2016)
Michie KA, Kwan AH, Tung CS, Guss JM, Trewhella J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
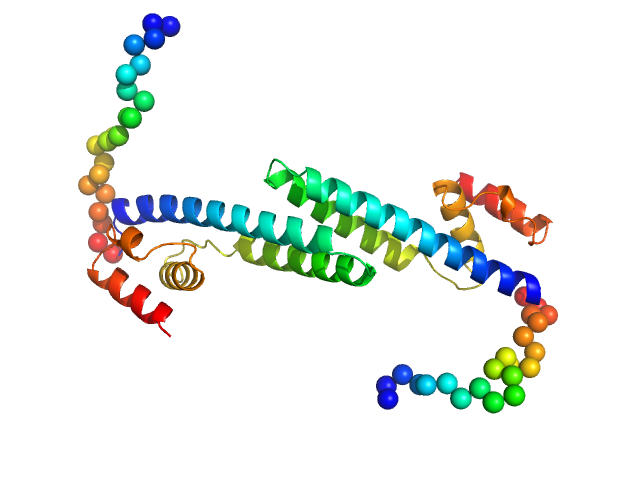
UniProt ID: P74103 (None-None) Fluorescence recovery protein
|
|
|
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
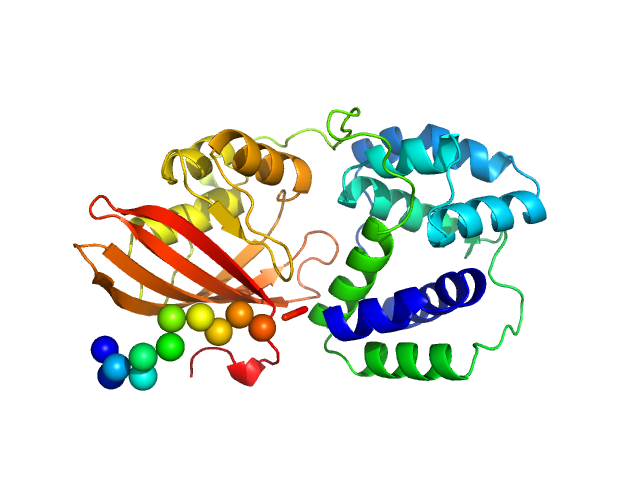
UniProt ID: P74102 (None-None) Orange carotenoid-binding protein
|
|
|
|
| Sample: |
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
57 |
nm3 |
|
|