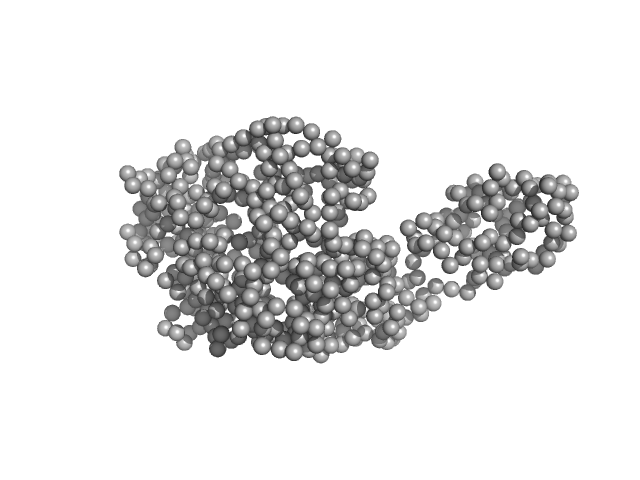
UniProt ID: P74103 (None-None) Fluorescence recovery protein
UniProt ID: P74102 (None-None) Orange carotenoid-binding protein
|
|
|
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
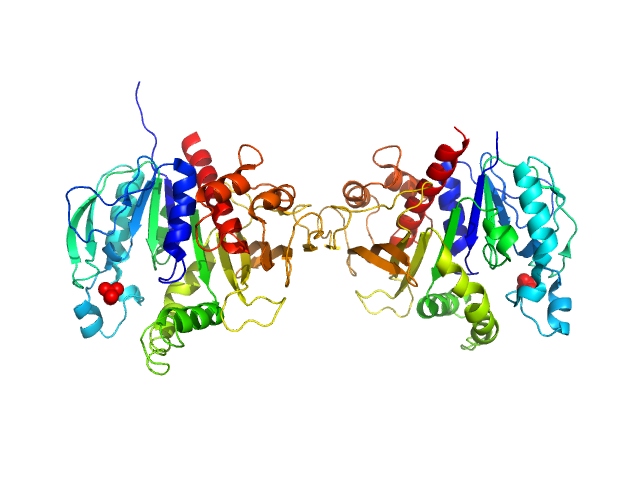
UniProt ID: P19824 (32-375) Phosphoribulokinase, chloroplastic
|
|
|
|
| Sample: |
Phosphoribulokinase, chloroplastic dimer, 78 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
Tris-HCl 50 mM 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Arabidopsis and Chlamydomonas phosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc Natl Acad Sci U S A 116(16):8048-8053 (2019)
Gurrieri L, Del Giudice A, Demitri N, Falini G, Pavel NV, Zaffagnini M, Polentarutti M, Crozet P, Marchand CH, Henri J, Trost P, Lemaire SD, Sparla F, Fermani S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
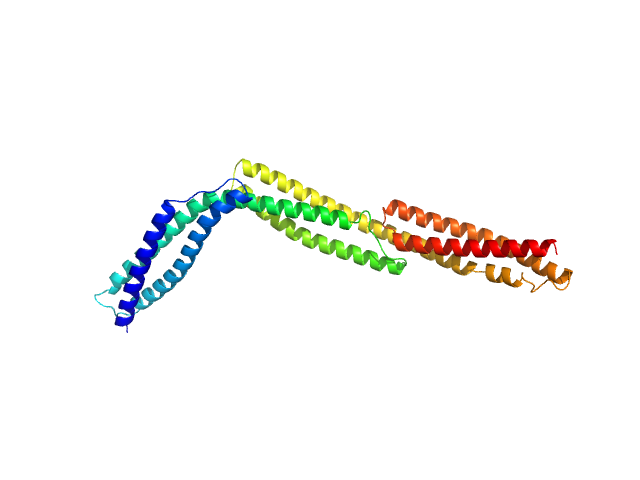
UniProt ID: P11532 (337-668) R1-3 human dystrophin fragment
|
|
|
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P11532 (338-668) R1-3 human dystrophin fragment
|
|
|

|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: P11532 (338-668) R1-3 human dystrophin fragment
|
|
|
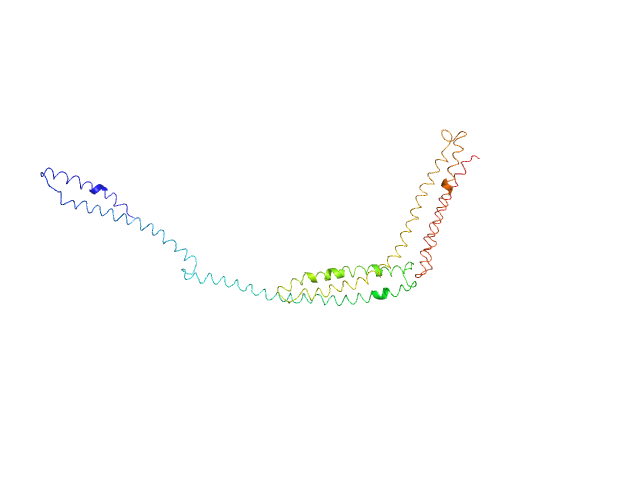
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: P06858 (29-475) Lipoprotein lipase
UniProt ID: None (None-None) Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1
|
|
|
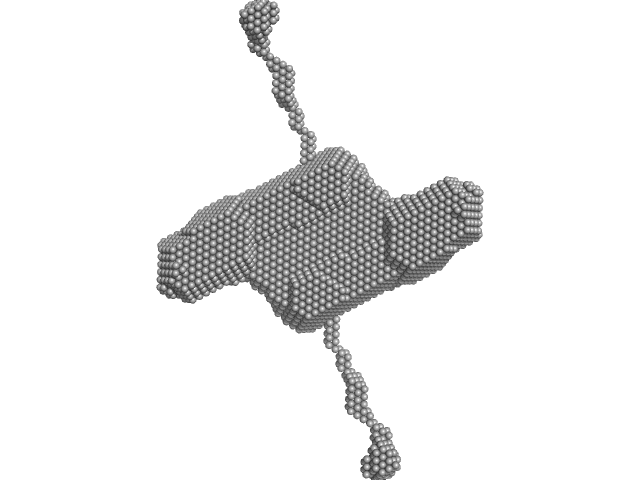
|
| Sample: |
Lipoprotein lipase dimer, 101 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
10 mM BisTris, 150 mM NaCl, 4 mM CaCl2, 10% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 2
|
Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis.
Proc Natl Acad Sci U S A (2018)
Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
319 |
nm3 |
|
|
UniProt ID: Q8TDZ2 (None-None) [F-actin]-monooxygenase MICAL1 (monomer)
|
|
|
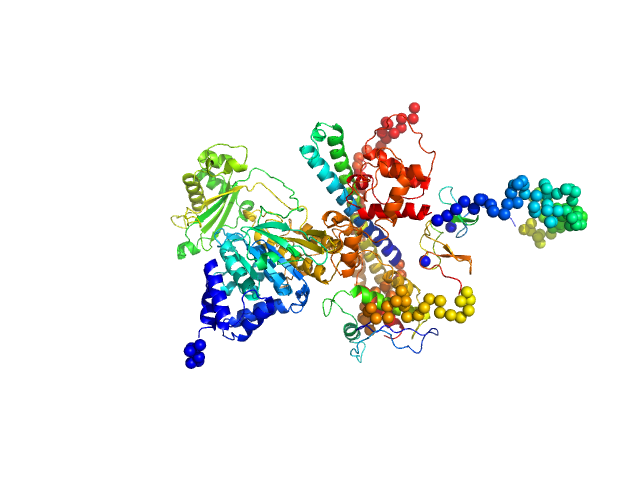
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, pH 7.5, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
UniProt ID: Q8TDZ2 (1-778) [F-actin]-monooxygenase MICAL1 (MoChLim)
|
|
|
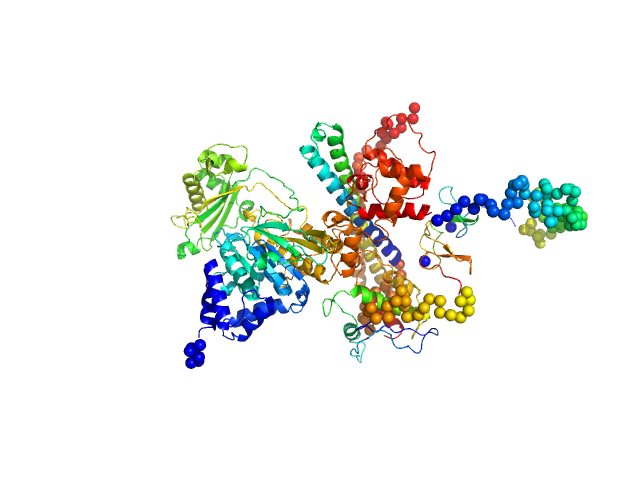
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoChLim) monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
UniProt ID: Q8TDZ2 (None-None) [F-actin]-monooxygenase MICAL1 (monomer)
UniProt ID: P61026 (1-175) Ras-related protein 8
|
|
|
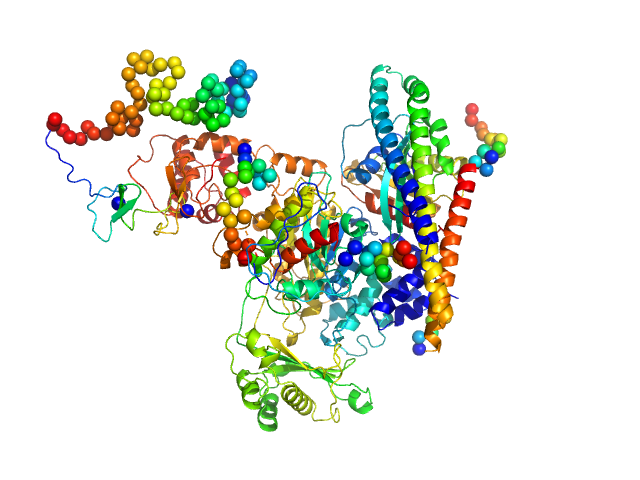
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
Ras-related protein 8 monomer, 20 kDa protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
UniProt ID: Q8TDZ2 (1-614) [F-actin]-monooxygenase MICAL1 (MoCh)
|
|
|
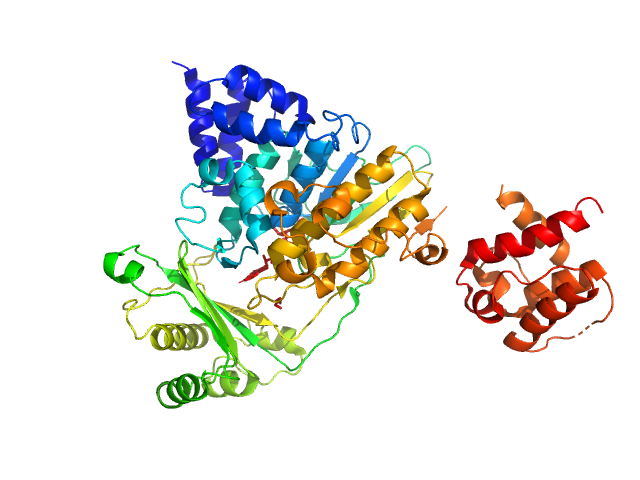
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoCh) monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 6
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|