UniProt ID: E7BBS4 (None-None) Insulin-like peptide 5
UniProt ID: Q09024 (None-None) Neural/ectodermal development factor IMP-L2
|
|
|
|
| Sample: |
Insulin-like peptide 5 monomer, 5 kDa Drosophila melanogaster protein
Neural/ectodermal development factor IMP-L2 monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 20
|
Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun 9(1):3860 (2018)
Roed NK, Viola CM, Kristensen O, Schluckebier G, Norrman M, Sajid W, Wade JD, Andersen AS, Kristensen C, Ganderton TR, Turkenburg JP, De Meyts P, Brzozowski AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
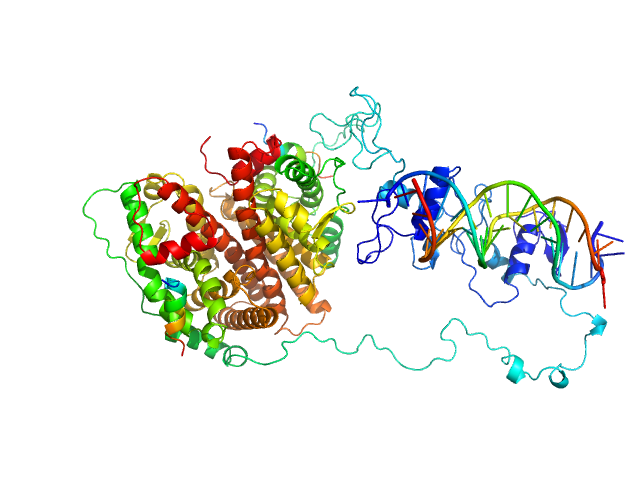
UniProt ID: P03372 (181-552) Estrogen receptor
UniProt ID: None (None-None) ERE1
UniProt ID: None (None-None) ERE2
UniProt ID: None (None-None) Estradiol
UniProt ID: None (None-None) hERa peptide1
UniProt ID: None (None-None) hERa peptide2
|
|
|
|
| Sample: |
Estrogen receptor dimer, 85 kDa protein
ERE1 monomer, 6 kDa Homo sapiens DNA
ERE2 monomer, 6 kDa Homo sapiens DNA
Estradiol dimer, 0 kDa
HERa peptide1 monomer, 2 kDa protein
HERa peptide2 monomer, 2 kDa protein
|
| Buffer: |
10 mM CHES (pH9.5), 125 mM NaCl, 5mM KCl, 4 mM MgCl2, 50 mM arginine, 50 mM glutamate, 5 mM TCEP, 5% glycerol, 10 µm Zn acetate, 10 µM estradiol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Aug 10
|
Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains.
Nat Commun 9(1):3520 (2018)
Huang W, Peng Y, Kiselar J, Zhao X, Albaqami A, Mendez D, Chen Y, Chakravarthy S, Gupta S, Ralston C, Kao HY, Chance MR, Yang S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
|
|
UniProt ID: P01012 (None-None) Ovalbumin
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
UniProt ID: Q6V115 (None-None) Ovalbumin (common quail)
|
|
|
|
| Sample: |
Ovalbumin (common quail) monomer, 42 kDa Coturnix coturnix protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
UniProt ID: P01012 (None-None) Ovalbumin
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
UniProt ID: P01012 (None-None) Ovalbumin
|
|
|

|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
0.16 M NaOH, pH: 13.2 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Feb 21
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
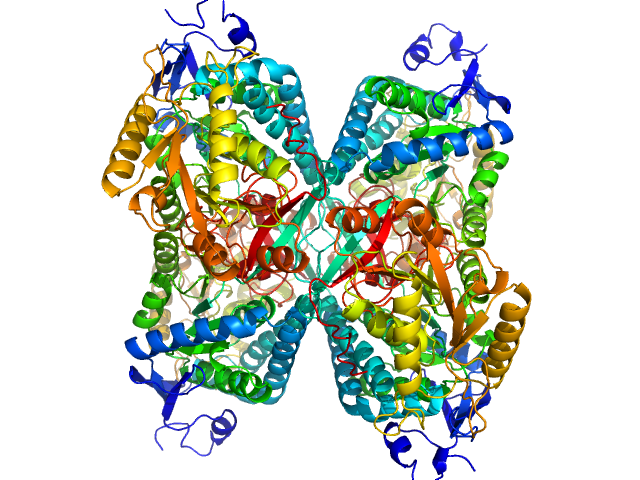
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
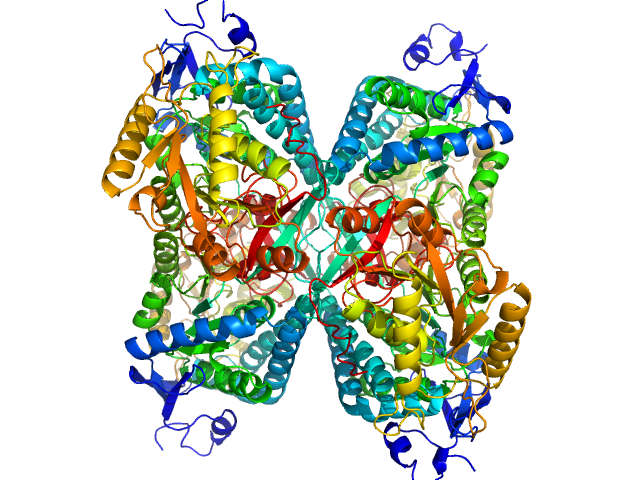
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
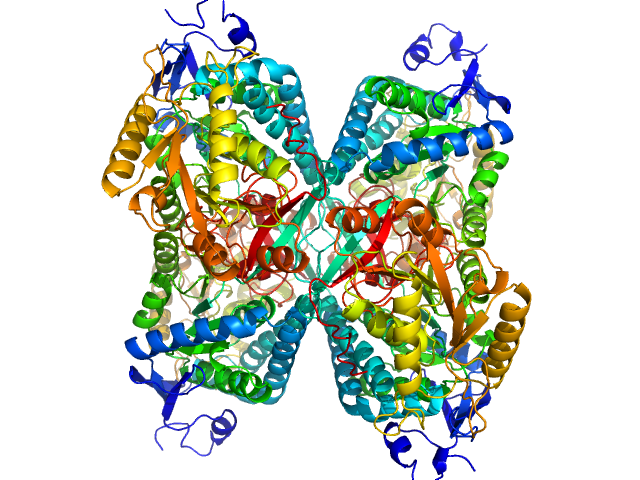
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
UniProt ID: P49419 (None-None) Alpha-aminoadipic semialdehyde dehydrogenase
|
|
|
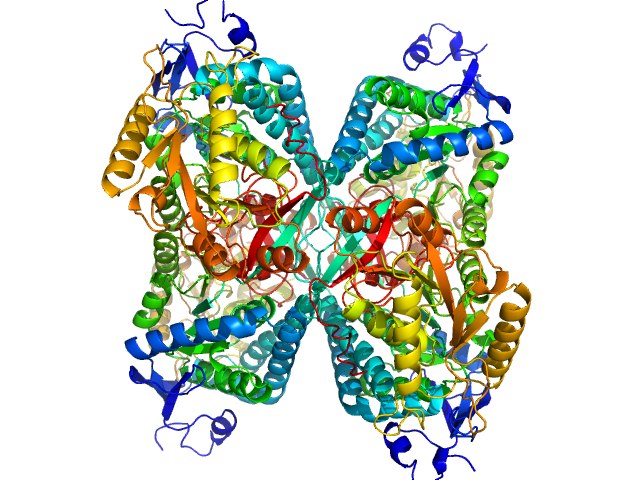
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|