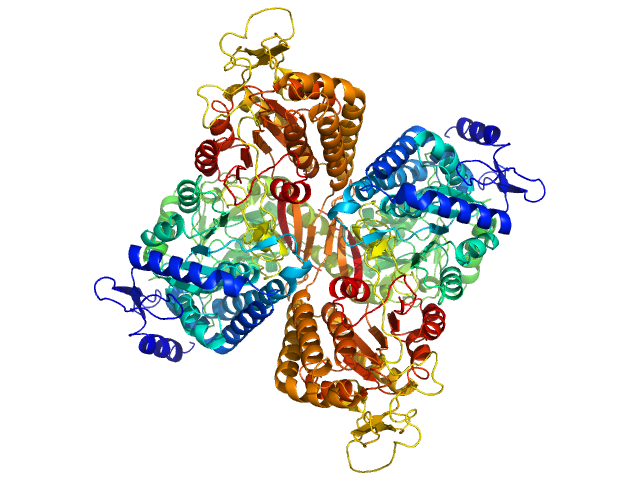
UniProt ID: Q8IZ83 (None-None) Aldehyde dehydrogenase family 16 member A1 from Homo sapiens
|
|
|
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
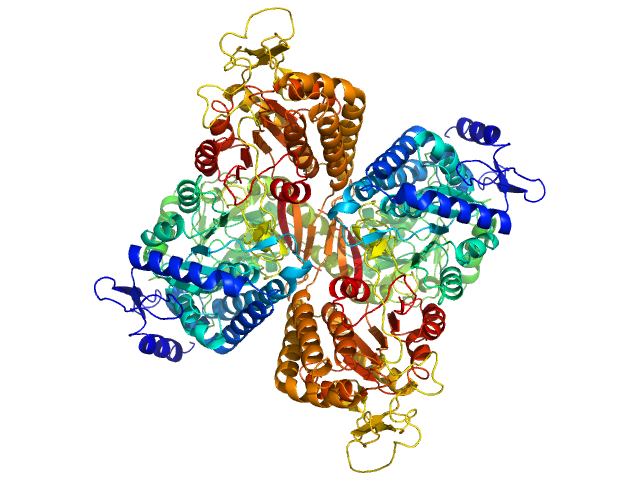
UniProt ID: Q8IZ83 (None-None) Aldehyde dehydrogenase family 16 member A1 from Homo sapiens
|
|
|
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
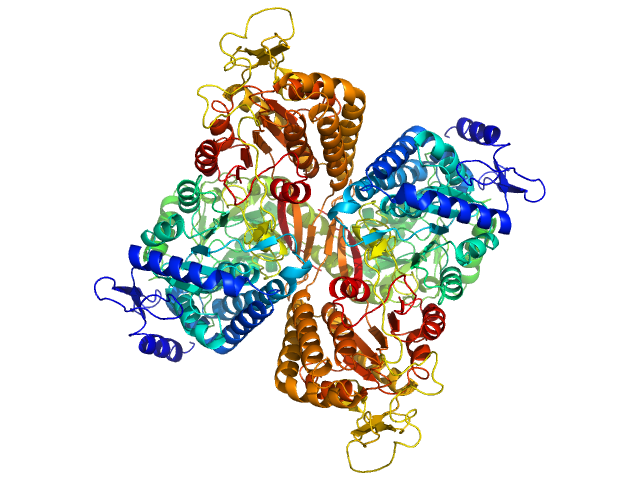
UniProt ID: Q8IZ83 (None-None) Aldehyde dehydrogenase family 16 member A1 from Homo sapiens
|
|
|
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
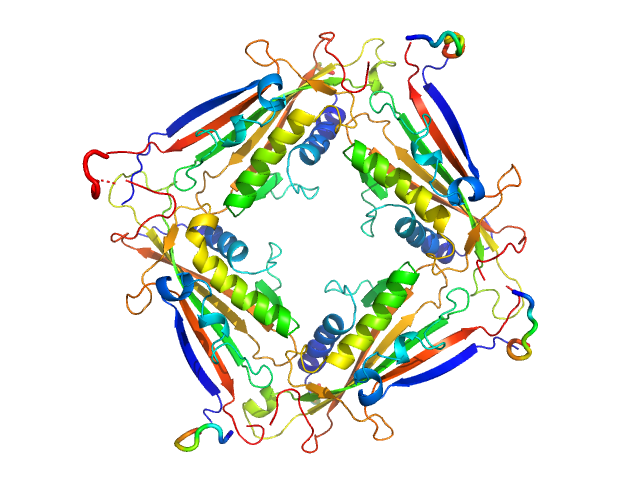
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
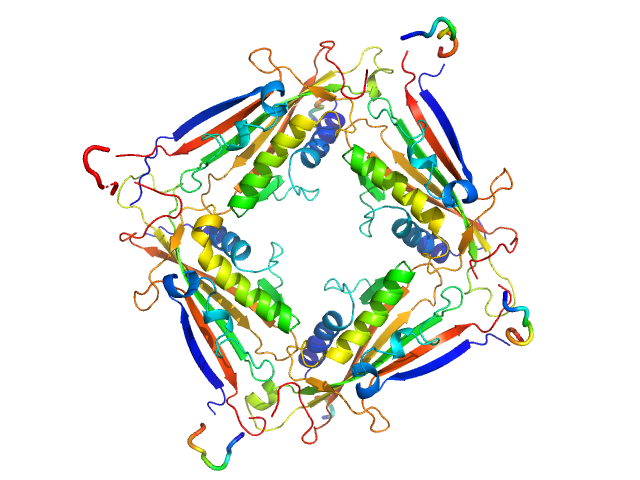
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
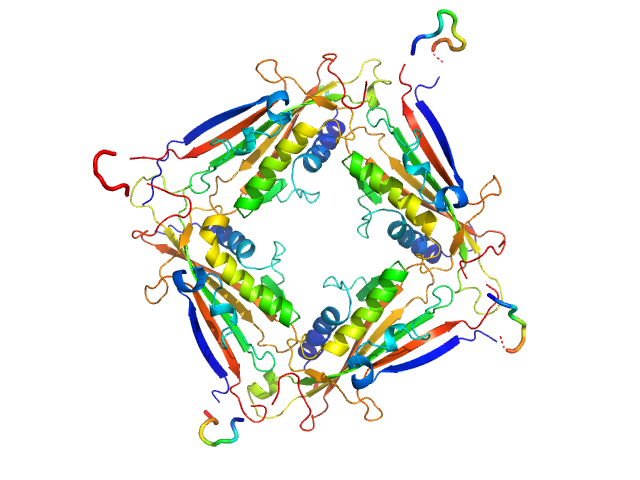
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 6.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
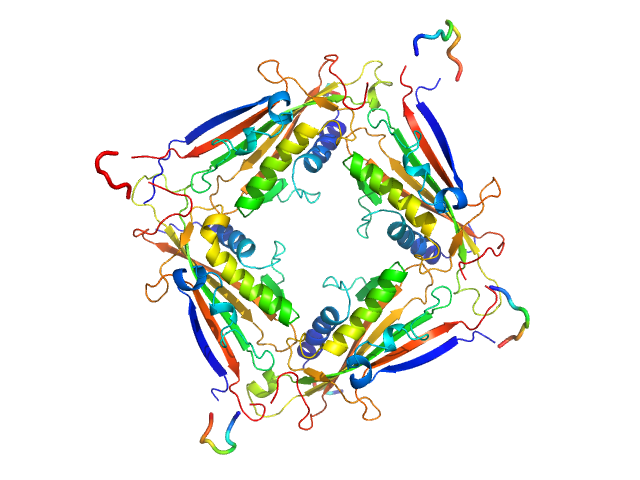
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 6.5 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
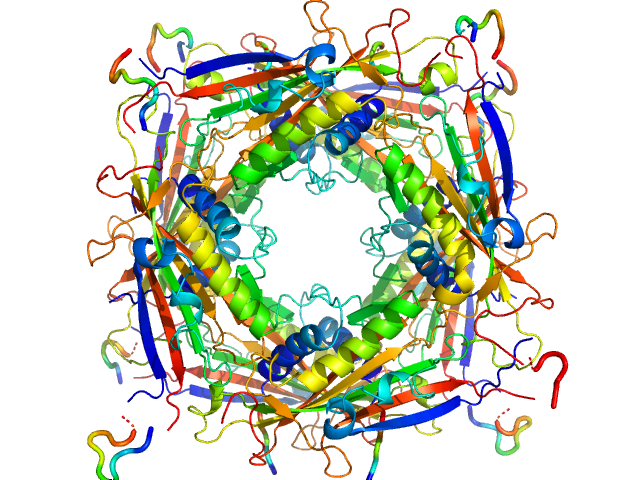
|
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
UniProt ID: Q8IZK6 (1-285) Transient receptor potential channel mucolipin 2
|
|
|
|
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
UniProt ID: P40699 (203-303) Surface presentation of antigens protein SpaO SpaO(SPOA2)
UniProt ID: P40699 (1-303) Surface presentation of antigens protein SpaO(SPOA1,2)
UniProt ID: P0CL45 (1-226) Oxygen-regulated invasion protein OrgB
UniProt ID: E8XL22 (1-431) ATP synthase InvC
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO SpaO(SPOA2) dimer, 25 kDa Salmonella enterica subsp. … protein
Surface presentation of antigens protein SpaO(SPOA1,2) monomer, 34 kDa Salmonella enterica subsp. … protein
Oxygen-regulated invasion protein OrgB dimer, 53 kDa Salmonella enterica subsp. … protein
ATP synthase InvC monomer, 48 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 12
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
22.7 |
nm |
| VolumePorod |
302 |
nm3 |
|
|