UniProt ID: O60828 (1-154) Polyglutamine-binding protein 1 p.Arg153Serfs*41
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 p.Arg153Serfs*41 dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2013 Feb 15
|
Frameshift PQBP-1 mutants K192Sfs*7 and R153Sfs*41 implicated in X-linked intellectual disability form stable dimers.
J Struct Biol (2019)
Rahman SK, Okazawa H, Chen YW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
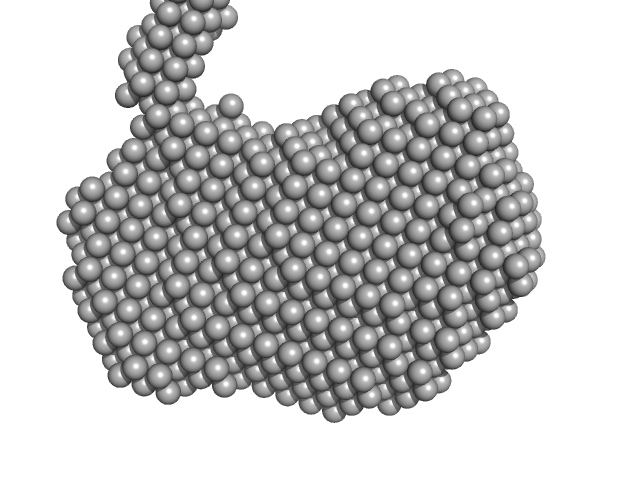
UniProt ID: E1B2U7 (1-476) LipAMS8
|
|
|
|
| Sample: |
LipAMS8 monomer, 50 kDa Pseudomonas sp. AMS8 protein
|
| Buffer: |
50 mM Tris HCl, 5 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Mar 6
|
Structural interpretations of a flexible cold-active AMS8 lipase by combining small-angle X-ray scattering and molecular dynamics simulation (SAXS-MD).
Int J Biol Macromol (2022)
Yaacob N, Kamonsutthipaijit N, Soontaranon S, Leow TC, Rahman RNZRA, Ali MSM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
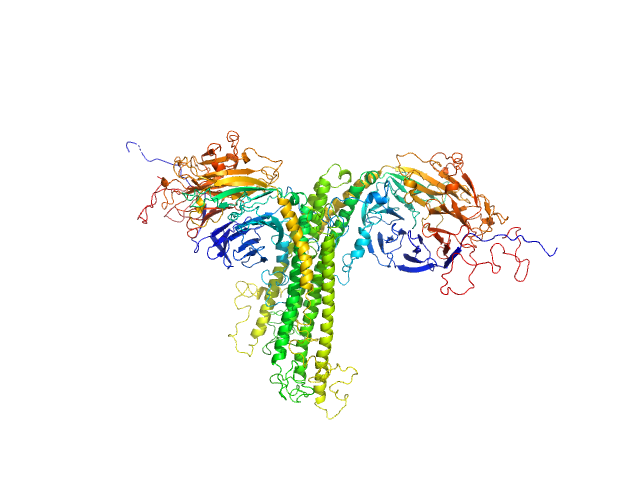
UniProt ID: O65418 (1-777) Anaphase Promoting Complex/Cyclosome Subunit 4
|
|
|
|
| Sample: |
Anaphase Promoting Complex/Cyclosome Subunit 4 dimer, 184 kDa Arabidopsis thaliana protein
|
| Buffer: |
10 mM Tris 150 mM NaCl 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Structural Characterisation of the Arabidopsis thaliana APC4
Steven De Gieter
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
259 |
nm3 |
|
|
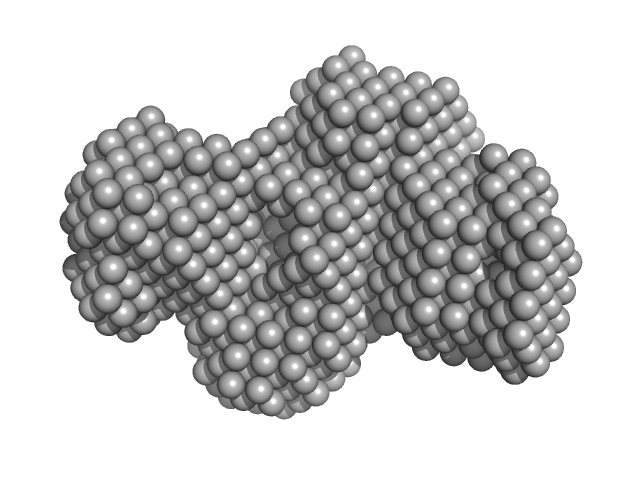
UniProt ID: P22914 (None-None) Gamma-crystallin S
|
|
|
|
| Sample: |
Gamma-crystallin S dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
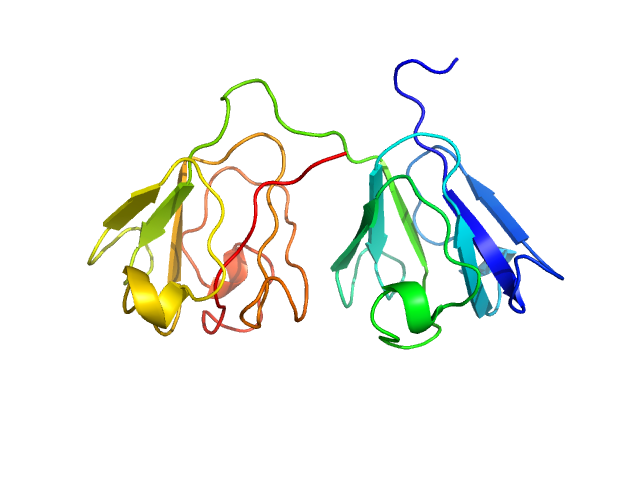
UniProt ID: P22914 (None-None) Gamma-crystallin S
|
|
|
|
| Sample: |
Gamma-crystallin S monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: Q9UGL1 (None-None) Lysine-specific demethylase 5B
|
|
|
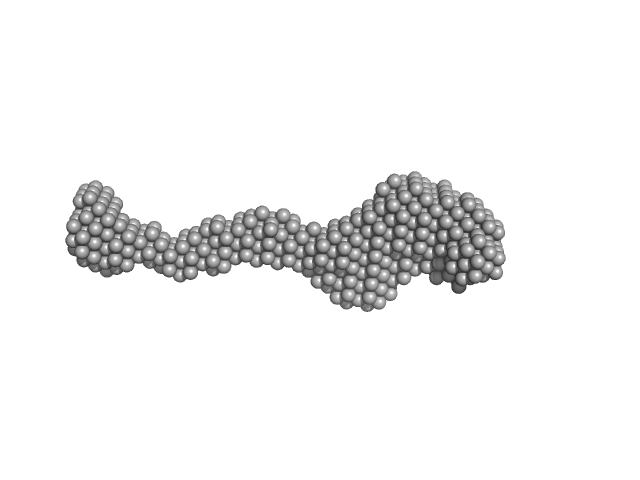
|
| Sample: |
Lysine-specific demethylase 5B monomer, 176 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 300 mM NaCl, 5% (v/v) glycerol, 1mM DTT, pH: 7.7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 24
|
Molecular architecture of the Jumonji C family histone demethylase KDM5B.
Sci Rep 9(1):4019 (2019)
Dorosz J, Kristensen LH, Aduri NG, Mirza O, Lousen R, Bucciarelli S, Mehta V, Sellés-Baiget S, Solbak SMØ, Bach A, Mesa P, Hernandez PA, Montoya G, Nguyen TTTN, Rand KD, Boesen T, Gajhede M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
26.9 |
nm |
|
|
UniProt ID: A0A0Q3EUQ3 (1-766) Aldehyde dehydrogenase 16 from Loktanella sp.
|
|
|
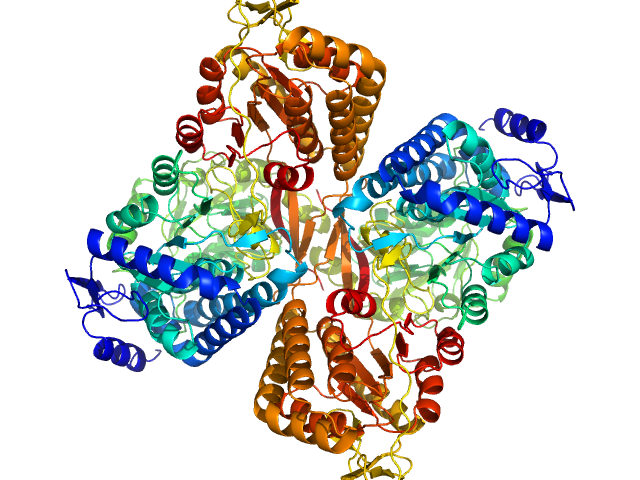
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
UniProt ID: A0A0Q3EUQ3 (1-766) Aldehyde dehydrogenase 16 from Loktanella sp.
|
|
|
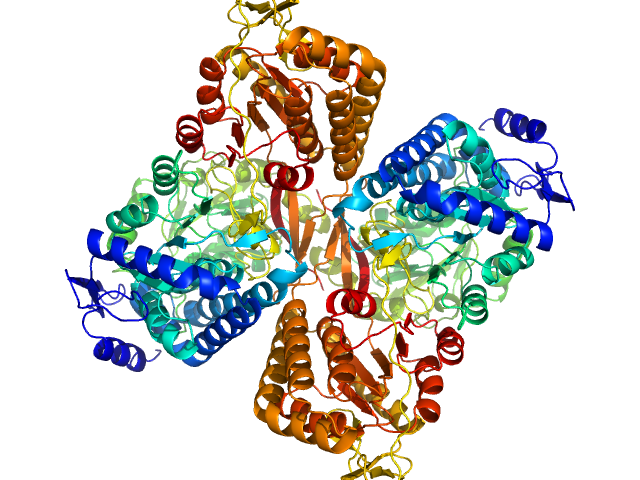
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
UniProt ID: A0A0Q3EUQ3 (1-766) Aldehyde dehydrogenase 16 from Loktanella sp.
|
|
|
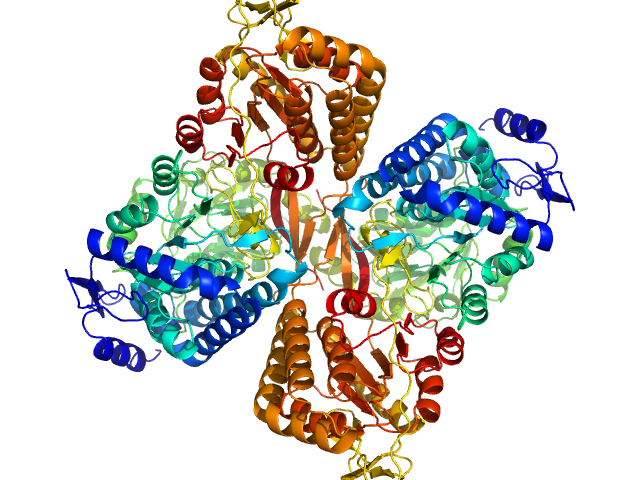
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
UniProt ID: A0A0Q3EUQ3 (1-766) Aldehyde dehydrogenase 16 from Loktanella sp.
|
|
|
|
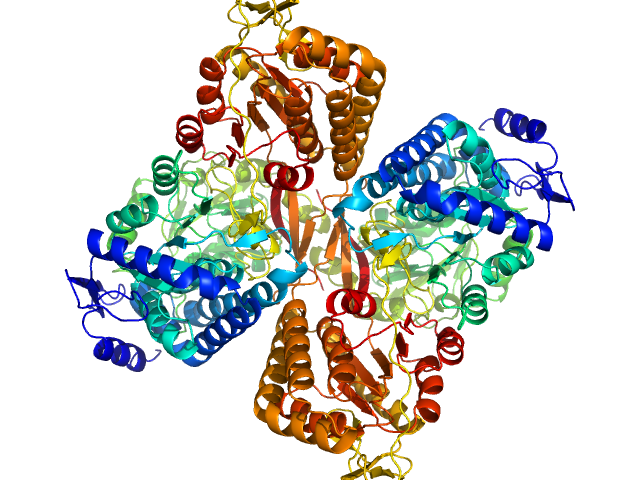
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
205 |
nm3 |
|
|