UniProt ID: P40699 (140-297) Surface presentation of antigens protein SpaO(SPOA1,2) C-terminus
|
|
|
|
| Sample: |
Surface presentation of antigens protein SpaO(SPOA1,2) C-terminus monomer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 24
|
Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes.
J Mol Biol 431(19):3787-3803 (2019)
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
|
|
UniProt ID: P23378 (45-1020) Glycine decarboxylase
|
|
|
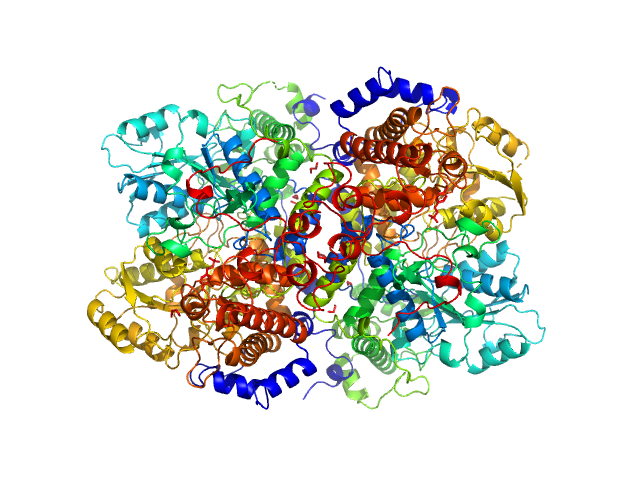
|
| Sample: |
Glycine decarboxylase dimer, 219 kDa Homo sapiens protein
|
| Buffer: |
10 mM TRIS pH 7.5, 100 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 12
|
Glycine decarboxylase (P-protein of glycine cleavage system)
Bart Van Laer
|
| RgGuinier |
3.9 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
UniProt ID: P23378 (45-1020) Glycine decarboxylase
|
|
|
|
| Sample: |
Glycine decarboxylase dimer, 219 kDa Homo sapiens protein
|
| Buffer: |
10 mM TRIS pH 7.5, 100 mM NaCl, 1 mM DTT , 100 mM glycine, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 12
|
Glycine decarboxylase (P-protein of glycine cleavage system)
Bart Van Laer
|
| RgGuinier |
4.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
UniProt ID: Q8VHV5 (90-231) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Myodes glareolus protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 25
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: Q8VHV5 (90-231) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Myodes glareolus protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, 0.001 mM CuSO4, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 25
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
UniProt ID: P23907 (94-234) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P23907 (94-234) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, 0.001 mM CuSO4, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: P23907 (94-234) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P23907 (94-234) Major prion protein
|
|
|
|
| Sample: |
Major prion protein monomer, 16 kDa Ovis aries protein
|
| Buffer: |
25 mM Ammonium Acetate, 250 mM NaCl, 0.001 mM CuSO4, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 4
|
Deciphering Copper Coordination in the Mammalian Prion Protein Amyloidogenic Domain
Biophysical Journal (2020)
Salzano G, Brennich M, Mancini G, Tran T, Legname G, D’Angelo P, Giachin G
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: Q88MI5 (1-91) Putative killer protein, Toxin GraT.
|
|
|
|
| Sample: |
Putative killer protein, Toxin GraT. monomer, 11 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Sep 21
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|