|
|
|
|
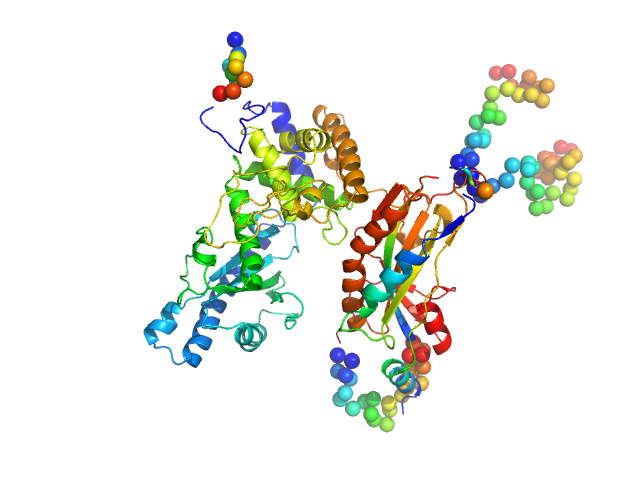
|
| Sample: |
NURS complex subunit red1 monomer, 8 kDa Schizosaccharomyces pombe (strain … protein
Poly(A) polymerase pla1 monomer, 65 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
20 mM HEPES, pH 7.5, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun 14(1):772 (2023)
Soni K, Sivadas A, Horvath A, Dobrev N, Hayashi R, Kiss L, Simon B, Wild K, Sinning I, Fischer T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
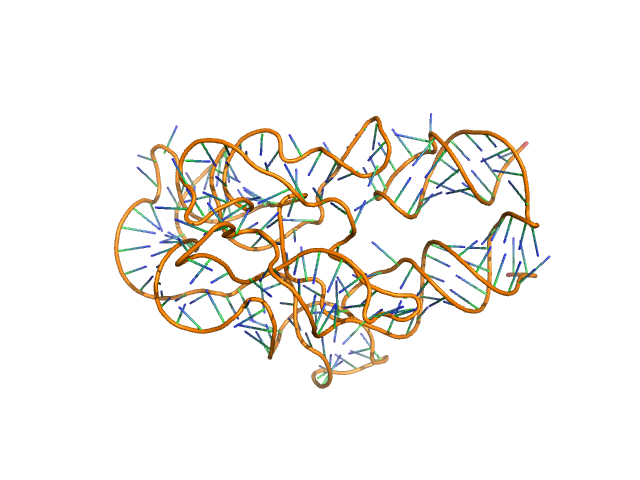
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.9 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
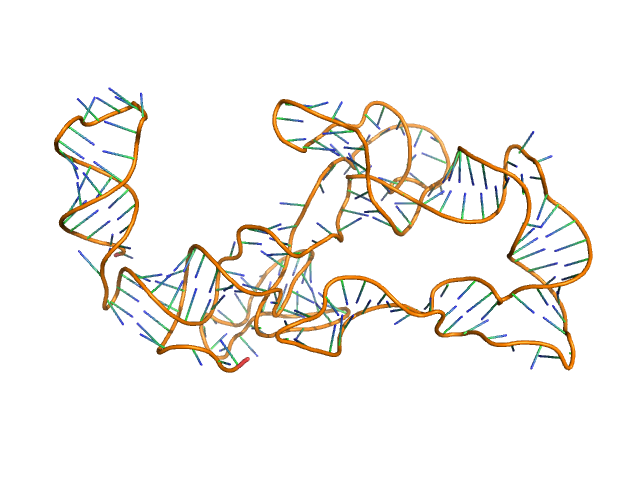
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, 4-18 μM coenzyme B12 ligand, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
P123 expressed protein dimer, 248 kDa Mycoplasma mobile (strain … protein
|
| Buffer: |
100 mM ammonium acetate, pH: 6.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Oct 25
|
Structure and Function of Gli123 Involved in Mycoplasma mobile Gliding.
J Bacteriol :e0034022 (2023)
Matsuike D, Tahara YO, Nonaka T, Wu HN, Hamaguchi T, Kudo H, Hayashi Y, Arai M, Miyata M
|
| RgGuinier |
8.2 |
nm |
| Dmax |
31.6 |
nm |
| VolumePorod |
436 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 22
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(CAA)6CA-RNA monomer, 6 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 22
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
(AG)10-24bp loading-RNA monomer, 15 kDa synthetic construct RNA
(AG)10-24bp duplex-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|