|
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
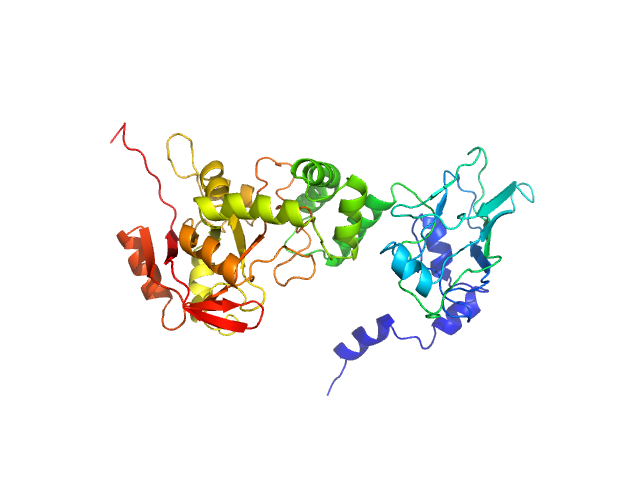
|
| Sample: |
NAD glycohydrolase monomer, 47 kDa Streptococcus pyogenes M1 … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 16
|
Structural basis underlying the synergism of NADase and SLO during group A Streptococcus infection.
Commun Biol 6(1):124 (2023)
Tsai WJ, Lai YH, Shi YA, Hammel M, Duff AP, Whitten AE, Wilde KL, Wu CM, Knott R, Jeng US, Kang CY, Hsu CY, Wu JL, Tsai PJ, Chiang-Ni C, Wu JJ, Lin YS, Liu CC, Senda T, Wang S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
103.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
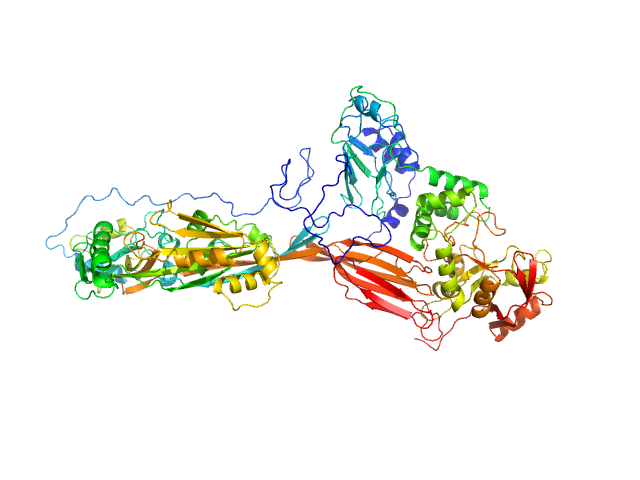
|
| Sample: |
NAD glycohydrolase monomer, 47 kDa Streptococcus pyogenes M1 … protein
Streptolysin O (T66M) monomer, 63 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Oct 16
|
Structural basis underlying the synergism of NADase and SLO during group A Streptococcus infection.
Commun Biol 6(1):124 (2023)
Tsai WJ, Lai YH, Shi YA, Hammel M, Duff AP, Whitten AE, Wilde KL, Wu CM, Knott R, Jeng US, Kang CY, Hsu CY, Wu JL, Tsai PJ, Chiang-Ni C, Wu JJ, Lin YS, Liu CC, Senda T, Wang S
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
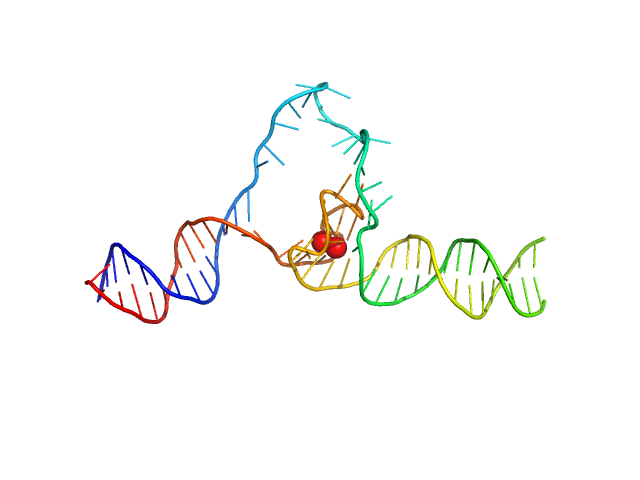
|
| Sample: |
Duplex-G4-duplex monomer, 29 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 16
|
Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 Å resolution with cryo-EM.
Nucleic Acids Res (2023)
Monsen RC, Chua EYD, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
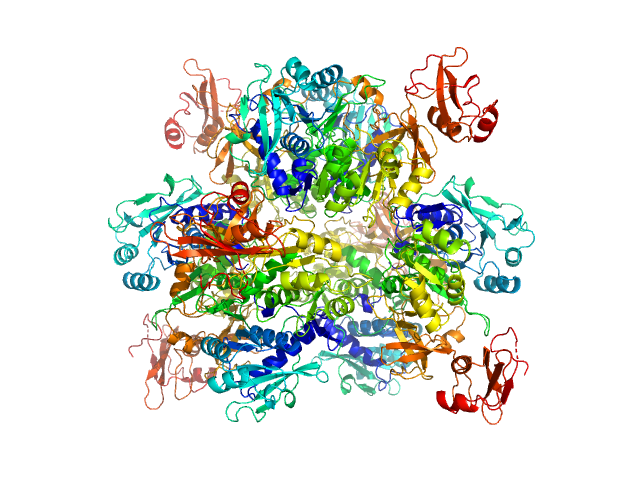
|
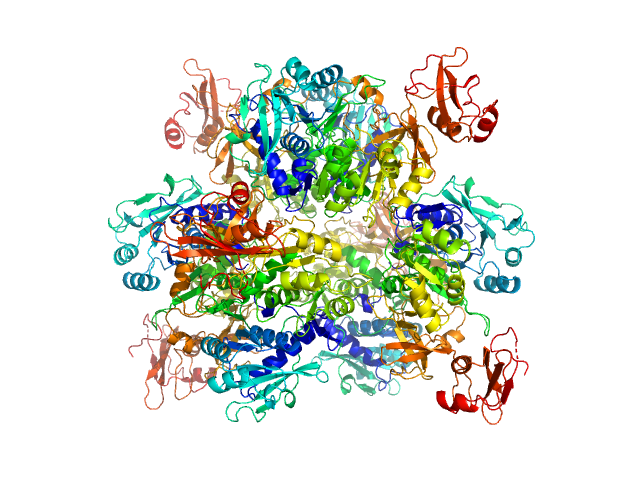
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
|
|
|
|
|
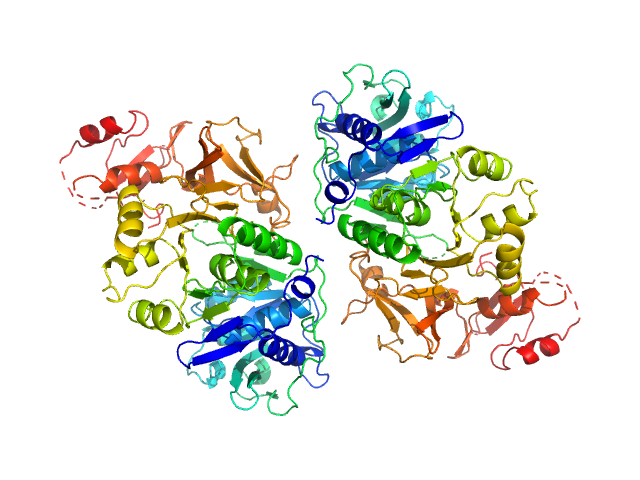
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
|
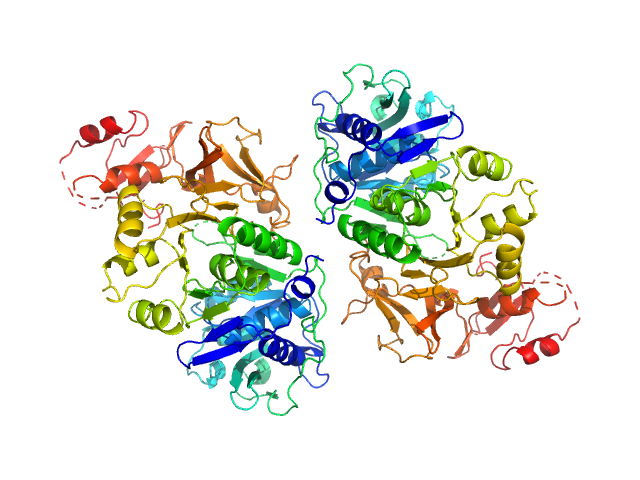
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 26 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 19
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|