|
|
|
|
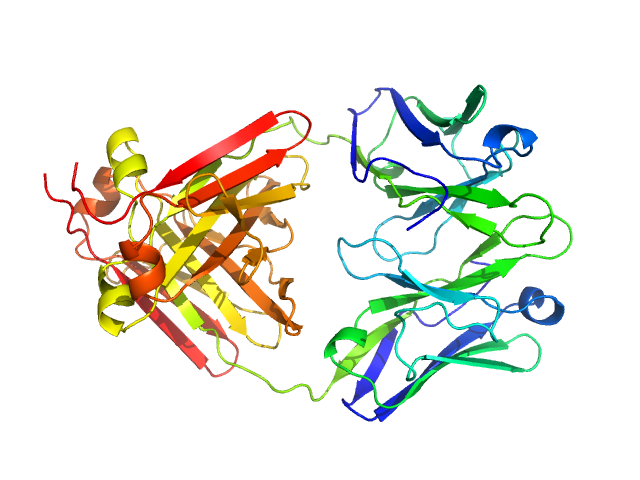
|
| Sample: |
Immunoglobulin light chain M7 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
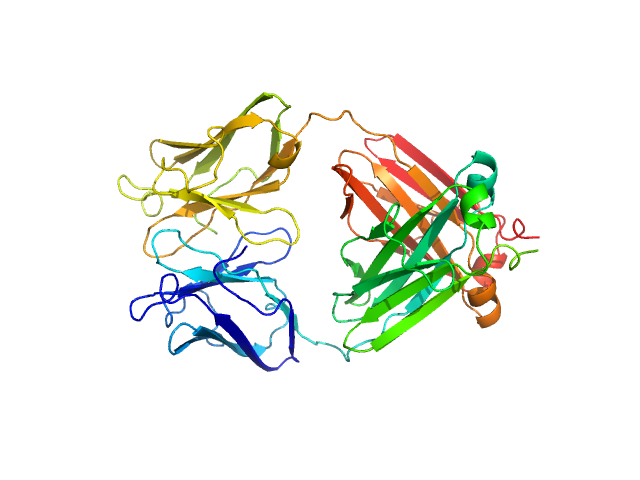
|
| Sample: |
Immunoglobulin light chain M10 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 17
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
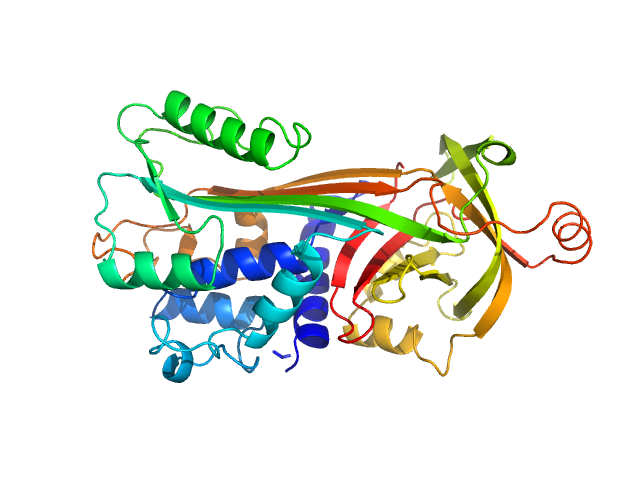
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
|
|
|
|
|
|
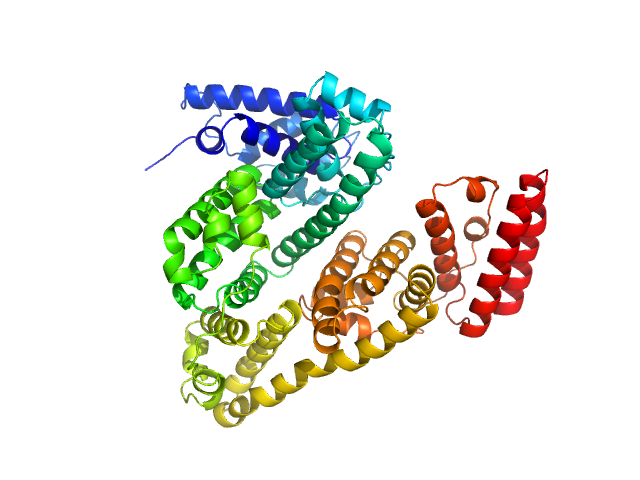
|
| Sample: |
Albumin (natural variant A214T) monomer, 66 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|
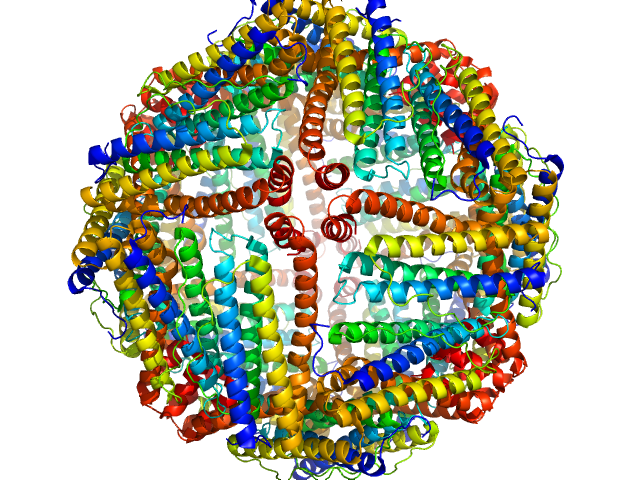
|
| Sample: |
Ferritin light chain 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.1 |
nm |
|
|
|
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiB hexamer, 71 kDa Synechococcus sp. (strain … protein
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiB hexamer, 71 kDa Synechococcus sp. (strain … protein
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
11.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
11.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
NAC domain-containing protein 13 dimer, 63 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NAC domain-containing protein 13 monomer, 13 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|