|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
7.0 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
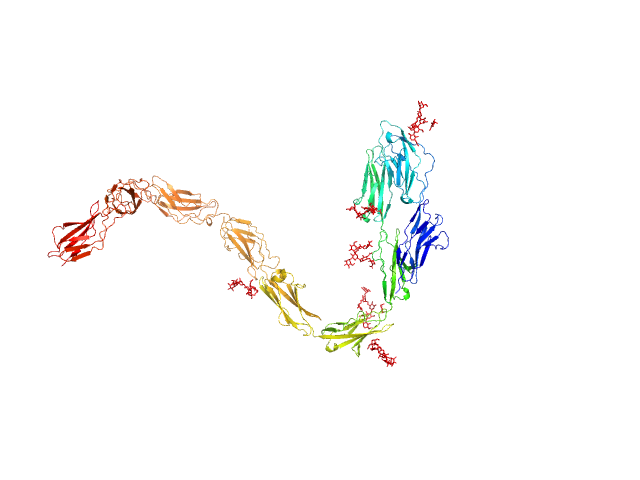
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl , 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 15
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA (Exonuclease III) (EXO III) (AP endonuclease VI) monomer, 33 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 15
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 31 kDa Mycobacterium tuberculosis protein
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 17
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
501 |
nm3 |
|
|
|
|
|
|

|
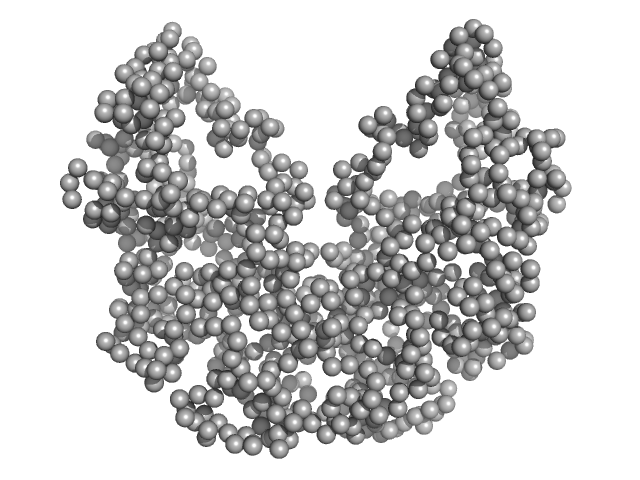
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 31 kDa Mycobacterium tuberculosis protein
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
DNA ligase A nicked DNA substrate dimer, 16 kDa DNA
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl , 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 17
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
|
|
|
|
|
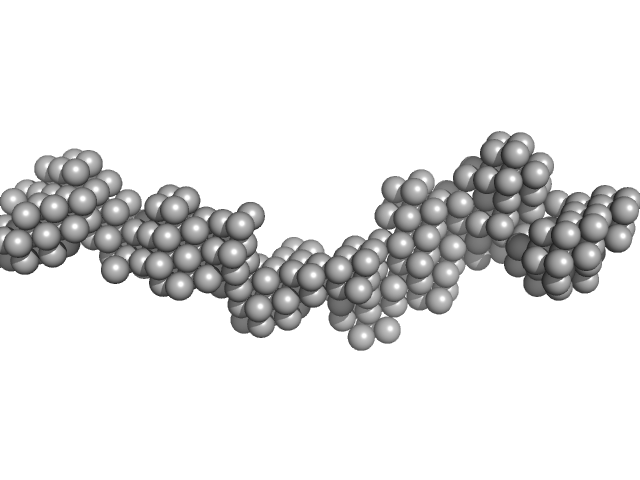
| Sample: |
Receptor-type tyrosine-protein phosphatase kappa monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
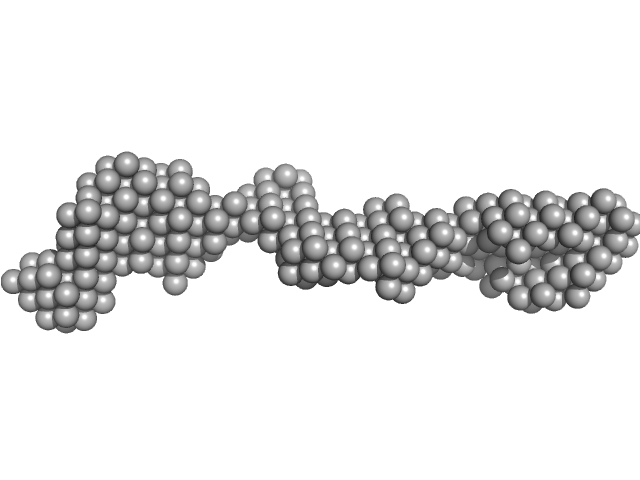
|
| Sample: |
Receptor-type tyrosine-protein phosphatase mu monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.2 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|