|
|
|
|
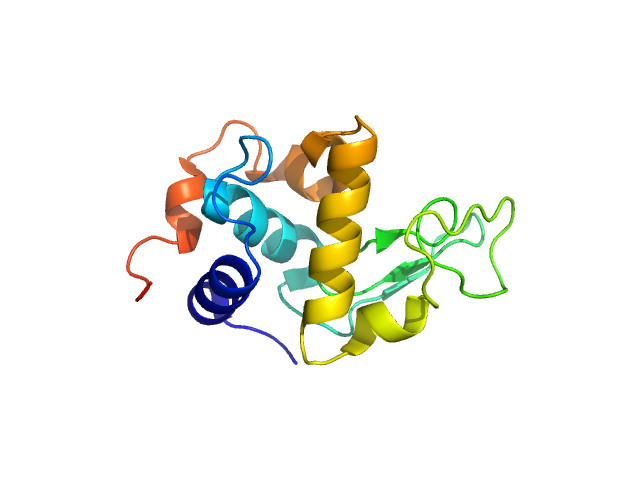
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
50 mM sodium citrate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.8 |
nm |
|
|
|
|
|
|
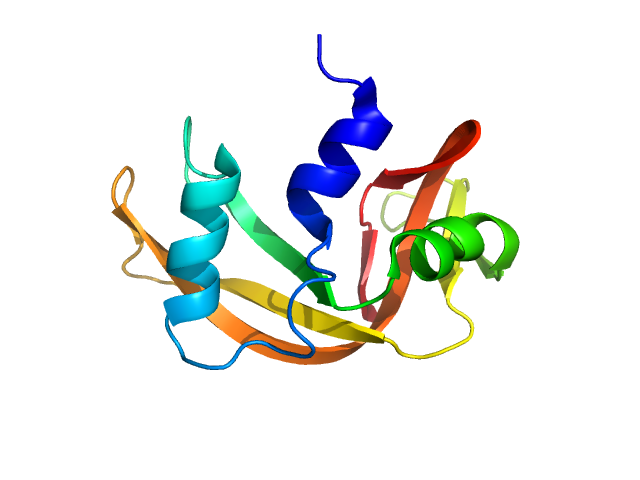
|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.1 |
nm |
|
|
|
|
|
|
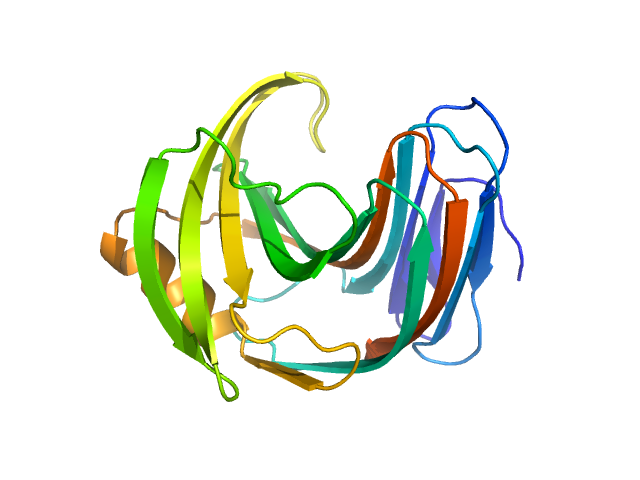
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
|
|
|
|
|
|
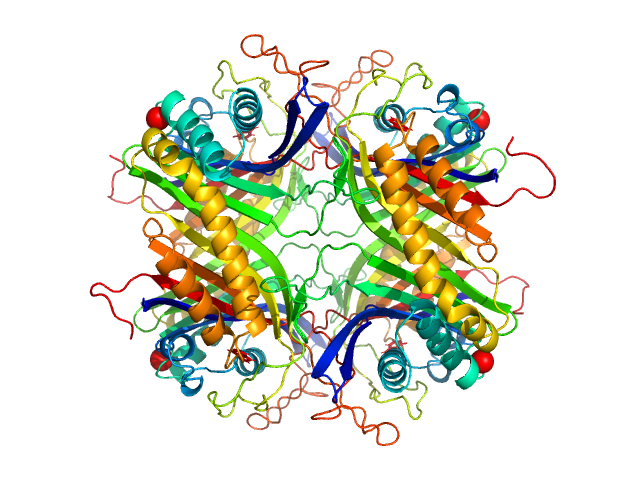
|
| Sample: |
Uricase tetramer, 136 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 24
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.1 |
nm |
|
|
|
|
|
|
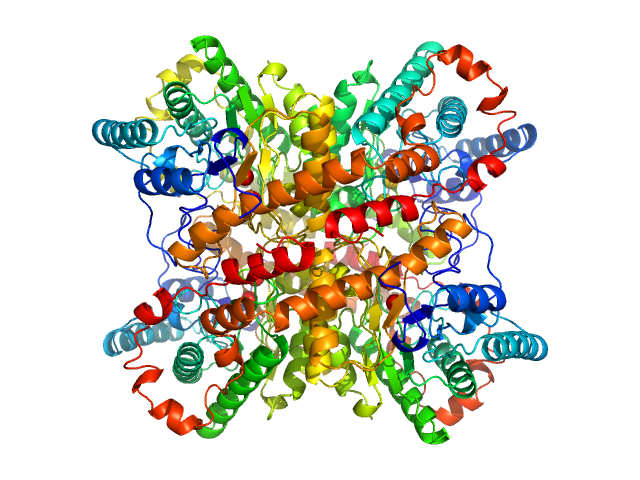
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 24
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
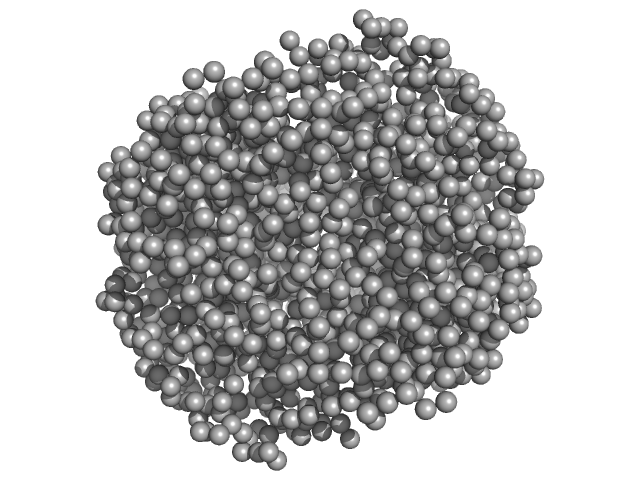
|
| Sample: |
HpcH/HpaI aldolase hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
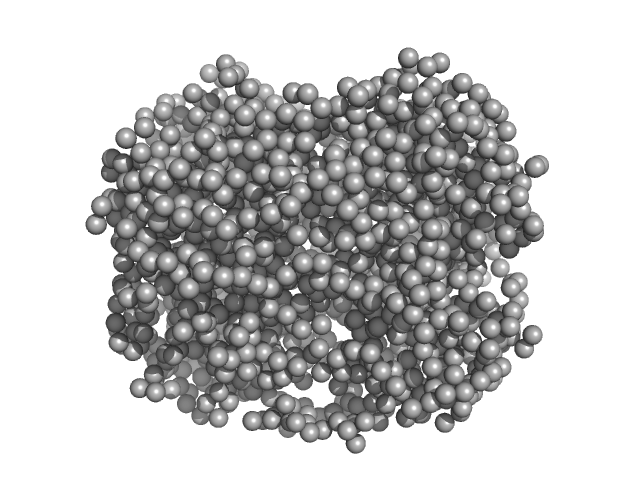
|
| Sample: |
HpcH/HpaI aldolase (S116A) hexamer, 165 kDa Sphingomonas wittichii (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
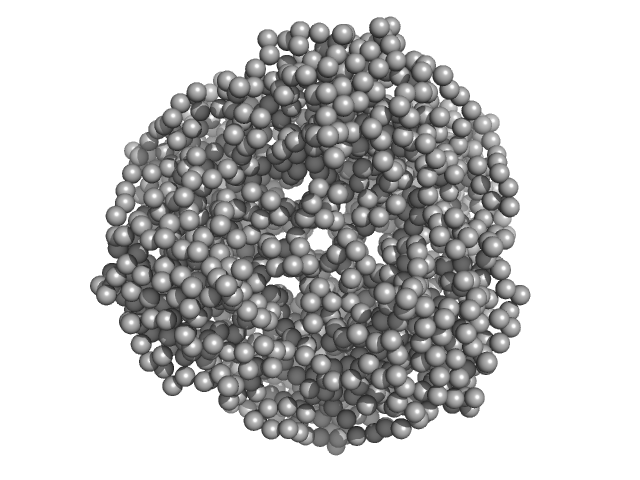
|
| Sample: |
HpcH/HpaI aldolase (S116C) hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
223 |
nm3 |
|
|