|
|
|
|
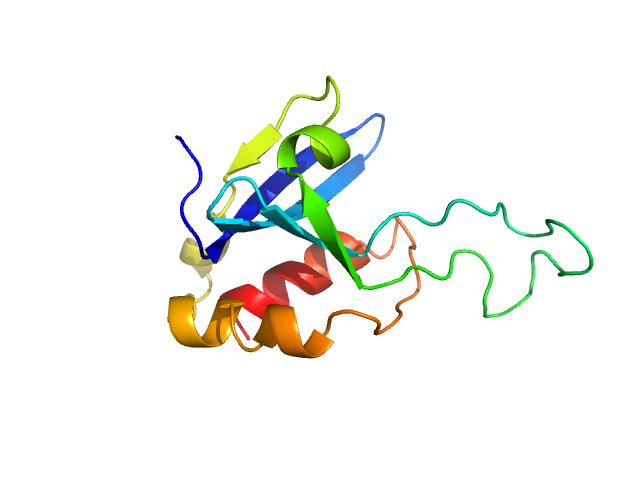
|
| Sample: |
Phospholipase A and acyltransferase 4 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TECP, 1% Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 11
|
Crystal structure of the phospholipase A and acyltransferase 4 (PLAAT4) catalytic domain.
J Struct Biol 214(4):107903 (2022)
Wehlin A, Cornaciu I, Marquez JA, Perrakis A, von Castelmur E
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable global transcription activator SNF2L2 (isoform 1) monomer, 16 kDa Homo sapiens protein
Von Hippel-Lindau disease tumor suppressor monomer, 19 kDa Homo sapiens protein
Elongin-B monomer, 12 kDa Homo sapiens protein
Elongin-C monomer, 11 kDa Homo sapiens protein
ACBI1 protac monomer, 1 kDa
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Aug 11
|
Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry.
Nat Commun 13(1):5884 (2022)
Dixon T, MacPherson D, Mostofian B, Dauzhenka T, Lotz S, McGee D, Shechter S, Shrestha UR, Wiewiora R, McDargh ZA, Pei F, Pal R, Ribeiro JV, Wilkerson T, Sachdeva V, Gao N, Jain S, Sparks S, Li Y, Vinitsky A, Zhang X, Razavi AM, Kolossváry I, Imbriglio J, Evdokimov A, Bergeron L, Zhou W, Adhikari J, Ruprecht B, Dickson A, Xu H, Sherman W, Izaguirre JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Von Hippel-Lindau disease tumor suppressor monomer, 19 kDa Homo sapiens protein
Elongin-B monomer, 12 kDa Homo sapiens protein
Elongin-C monomer, 11 kDa Homo sapiens protein
ACBI1 protac monomer, 1 kDa
Probable global transcription activator SNF2L2 (isoform 2) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Aug 11
|
Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry.
Nat Commun 13(1):5884 (2022)
Dixon T, MacPherson D, Mostofian B, Dauzhenka T, Lotz S, McGee D, Shechter S, Shrestha UR, Wiewiora R, McDargh ZA, Pei F, Pal R, Ribeiro JV, Wilkerson T, Sachdeva V, Gao N, Jain S, Sparks S, Li Y, Vinitsky A, Zhang X, Razavi AM, Kolossváry I, Imbriglio J, Evdokimov A, Bergeron L, Zhou W, Adhikari J, Ruprecht B, Dickson A, Xu H, Sherman W, Izaguirre JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
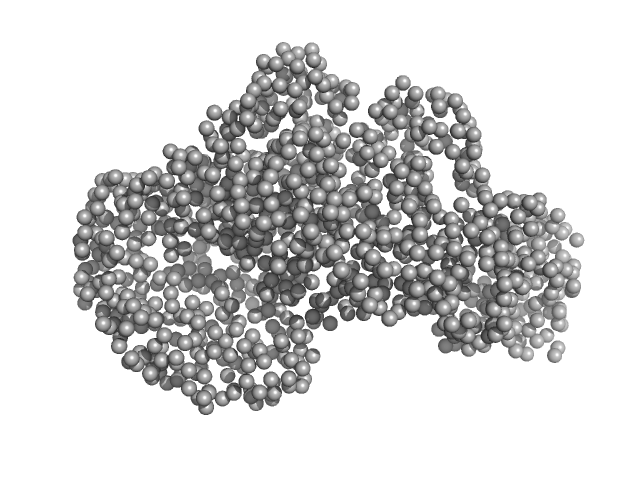
|
| Sample: |
Piwi domain protein monomer, 53 kDa Geobacter sulfurreducens (strain … protein
Sir2 superfamily protein monomer, 68 kDa Geobacter sulfurreducens (strain … protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion
Nature Microbiology (2022)
Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filaggrin-like protein monomer, 20 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 22
|
Intrinsically disordered plant protein PARCL co-localizes with RNA in phase-separated condensates whose formation can be regulated by mutating the PLD
Journal of Biological Chemistry :102631 (2022)
Ostendorp A, Ostendorp S, Zhou Y, Chaudron Z, Wolffram L, Rombi K, von Pein L, Falke S, Jeffries C, Svergun D, Betzel C, Morris R, Kragler F, Kehr J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filaggrin-like protein monomer, 20 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 22
|
Intrinsically disordered plant protein PARCL co-localizes with RNA in phase-separated condensates whose formation can be regulated by mutating the PLD
Journal of Biological Chemistry :102631 (2022)
Ostendorp A, Ostendorp S, Zhou Y, Chaudron Z, Wolffram L, Rombi K, von Pein L, Falke S, Jeffries C, Svergun D, Betzel C, Morris R, Kragler F, Kehr J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
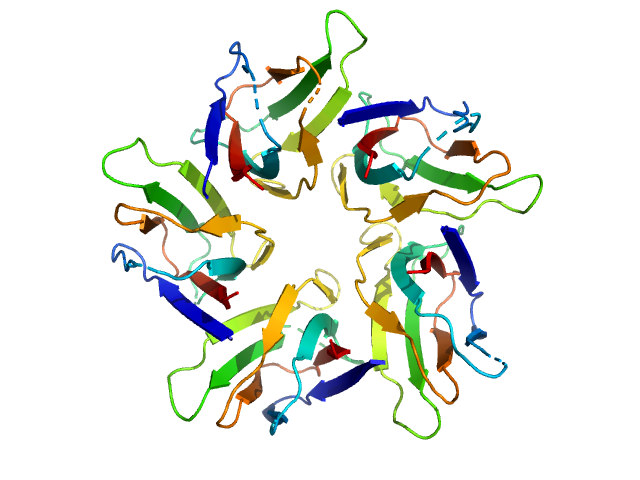
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 58 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 16
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
141 |
nm3 |
|
|