|
|
|
|
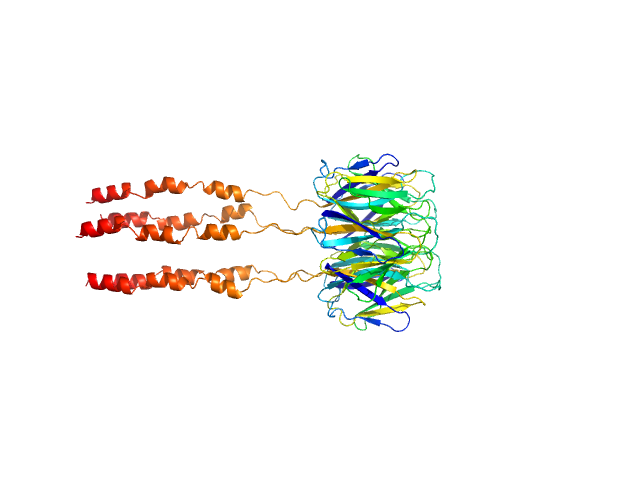
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
164 |
nm3 |
|
|
|
|
|
|
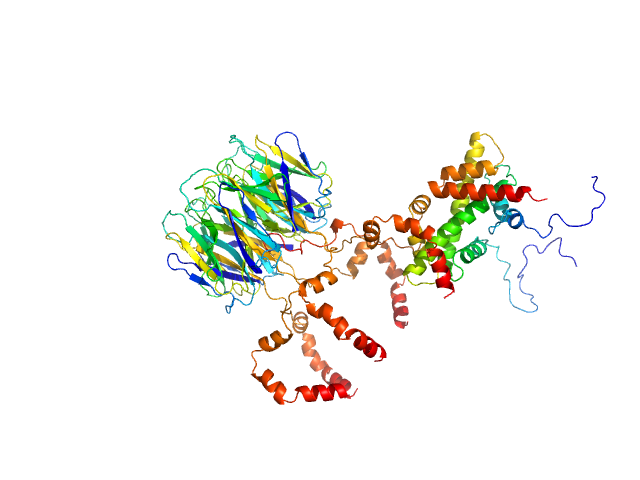
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
Histone H2A type 1-B/E monomer, 14 kDa Homo sapiens protein
Histone H2B type 1-C/E/F/G/I monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|
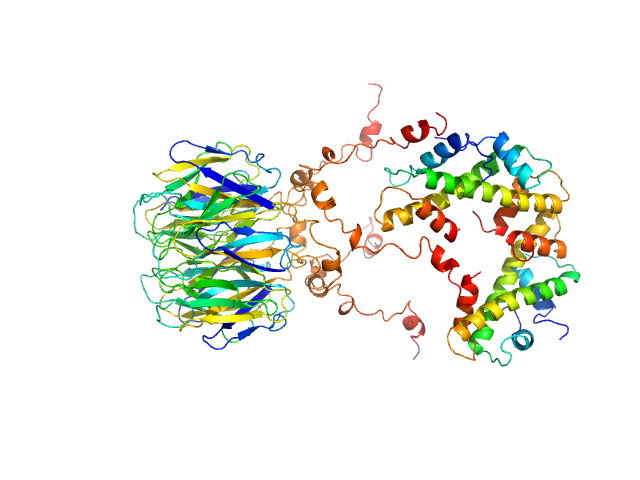
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 81 kDa Arabidopsis thaliana protein
Histone H3.1 dimer, 31 kDa Homo sapiens protein
Histone H4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|
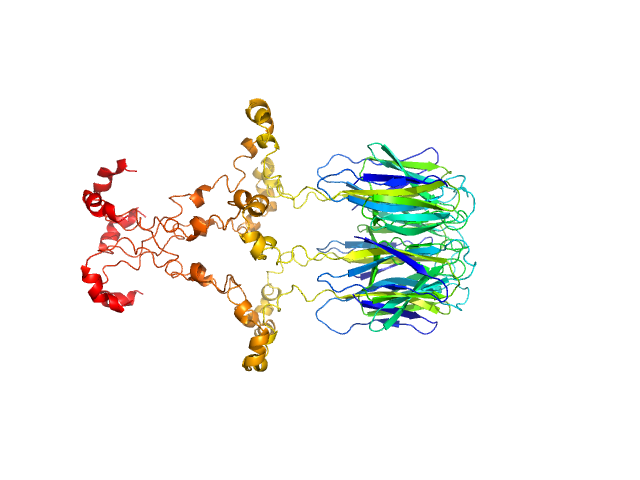
|
| Sample: |
Peptidyl-prolyl cis-trans isomerase FKBP43 pentamer, 96 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 28
|
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Biochim Biophys Acta Gene Regul Mech 1865(7):194872 (2022)
Singh AK, Saharan K, Baral S, Vasudevan D
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
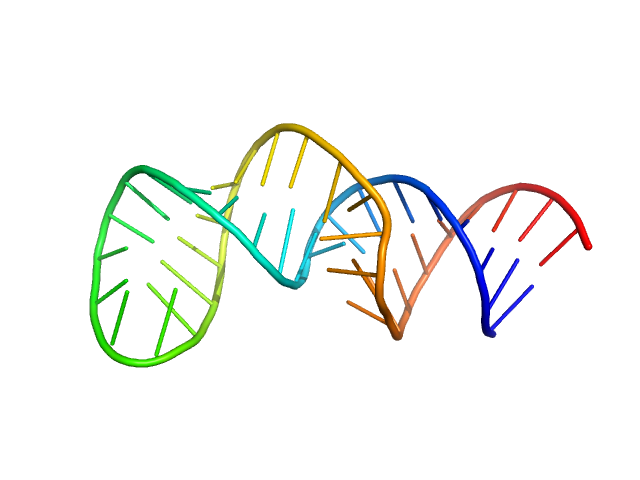
| Sample: |
The second exon splicing silencer 2p monomer, 14 kDa RNA
|
| Buffer: |
20 mM Bis-Tris, 20 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 1
|
Encoded Conformational Dynamics of the HIV Splice Site A3 Regulatory Locus: Implications for Differential Binding of hnRNP Splicing Auxiliary Factors.
J Mol Biol 434(18):167728 (2022)
Chiu LY, Emery A, Jain N, Sugarman A, Kendrick N, Luo L, Ford W, Swanstrom R, Tolbert BS
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-binding protein monomer, 13 kDa Mesorhizobium loti protein
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Crystallographic and X-ray scattering study of RdfS, a recombination directionality factor from an integrative and conjugative element
Acta Crystallographica Section D Structural Biology 78(10) (2022)
Verdonk C, Marshall A, Ramsay J, Bond C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|

|
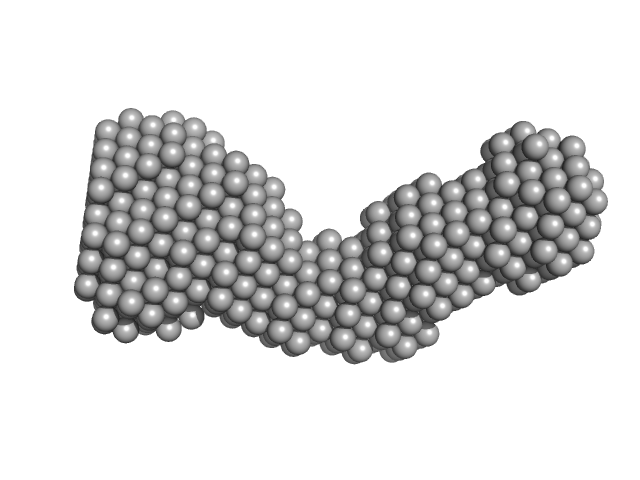
| Sample: |
Chromatin assembly factor 1 subunit A monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 200 mM NaCl, 0.1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Oct 28
|
Unorthodox PCNA Binding by Chromatin Assembly Factor 1
International Journal of Molecular Sciences 23(19):11099 (2022)
Gopinathan Nair A, Rabas N, Lejon S, Homiski C, Osborne M, Cyr N, Sverzhinsky A, Melendy T, Pascal J, Laue E, Borden K, Omichinski J, Verreault A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sperm acrosome membrane-associated protein 6 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Dec 11
|
SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol 5(1):984 (2022)
Vance TDR, Yip P, Jiménez E, Li S, Gawol D, Byrnes J, Usón I, Ziyyat A, Lee JE
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone-lysine N-methyltransferase 2A monomer, 25 kDa Homo sapiens protein
WD repeat-containing protein 5 monomer, 37 kDa Homo sapiens protein
Retinoblastoma-binding protein 5 monomer, 59 kDa Homo sapiens protein
Set1/Ash2 histone methyltransferase complex subunit ASH2 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TECP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
DPY30 acts as an ASH2L-specific stabilizer to stimulate the enzyme activity of MLL family methyltransferases on different substrates.
iScience 25(9):104948 (2022)
Zhao L, Huang N, Mencius J, Li Y, Xu Y, Zheng Y, He W, Li N, Zheng J, Zhuang M, Quan S, Chen Y
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone-lysine N-methyltransferase 2A monomer, 25 kDa Homo sapiens protein
WD repeat-containing protein 5 monomer, 37 kDa Homo sapiens protein
Retinoblastoma-binding protein 5 monomer, 59 kDa Homo sapiens protein
Set1/Ash2 histone methyltransferase complex subunit ASH2 monomer, 60 kDa Homo sapiens protein
Protein dpy-30 homolog dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TECP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
DPY30 acts as an ASH2L-specific stabilizer to stimulate the enzyme activity of MLL family methyltransferases on different substrates.
iScience 25(9):104948 (2022)
Zhao L, Huang N, Mencius J, Li Y, Xu Y, Zheng Y, He W, Li N, Zheng J, Zhuang M, Quan S, Chen Y
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
411 |
nm3 |
|
|